-
PDF
- Split View
-
Views
-
Cite
Cite
Damian Smedley, Paul Schofield, Chao-Kung Chen, Vassilis Aidinis, Chrysanthi Ainali, Jonathan Bard, Rudi Balling, Ewan Birney, Andrew Blake, Erik Bongcam-Rudloff, Anthony J. Brookes, Gianni Cesareni, Christina Chandras, Janan Eppig, Paul Flicek, Georgios Gkoutos, Simon Greenaway, Michael Gruenberger, Jean-Karim Hériché, Andrew Lyall, Ann-Marie Mallon, Dawn Muddyman, Florian Reisinger, Martin Ringwald, Nadia Rosenthal, Klaus Schughart, Morris Swertz, Gudmundur A. Thorisson, Michael Zouberakis, John M. Hancock, Finding and sharing: new approaches to registries of databases and services for the biomedical sciences, Database, Volume 2010, 2010, baq014, https://doi.org/10.1093/database/baq014
Close - Share Icon Share
Abstract
The recent explosion of biological data and the concomitant proliferation of distributed databases make it challenging for biologists and bioinformaticians to discover the best data resources for their needs, and the most efficient way to access and use them. Despite a rapid acceleration in uptake of syntactic and semantic standards for interoperability, it is still difficult for users to find which databases support the standards and interfaces that they need. To solve these problems, several groups are developing registries of databases that capture key metadata describing the biological scope, utility, accessibility, ease-of-use and existence of web services allowing interoperability between resources. Here, we describe some of these initiatives including a novel formalism, the Database Description Framework, for describing database operations and functionality and encouraging good database practise. We expect such approaches will result in improved discovery, uptake and utilization of data resources.
Database URL:http://www.casimir.org.uk/casimir_ddf
Biologists currently face a daunting challenge when trying to discover which of the multitude of computational and data resources to use in analysing their results and developing their hypotheses. The basic task of identifying appropriate online resources in a research field is non-trivial and typically involves ad hoc Internet trawling, recommendations from colleagues or literature searching. This is then followed by the more complex task of establishing whether the resource is relevant, reliable, well curated, and maintained. If programmatic access is required, discovering whether this exists and how to utilize it is another challenge. As time is short, most researchers often end up using familiar resources, which are not always the best or most relevant, while the developers and funders of under-utilized but valuable resources essentially waste time and money. What is required is a solution that helps to maximize the usefulness of each resource to the overall community. At present, approaches are being developed to construct two types of registry. One type, ‘databases of databases’, deal with describing the contents and other metadata about databases. The other type, web service registries, deal with the explicit description of services available at particular sites (not always databases). We present the two areas separately, but ultimately we expect solutions to arrive that merge the two approaches.
Registries of databases
Comprehensive, top-level registries of biological resources are currently provided by the Nucleic Acids Research Molecular Biology Database Collection,1 the BioMedCentral Catalog of Databases on the Web (http://databases.biomedcentral.com) and the Bioinformatics.ca Links Directory.2 However, they do not collect extensive metadata beyond a brief description of the resource and URL, can only be browsed by the category each registry has assigned or searched by the resource name, and lack much of the detailed information that the community requires. A number of projects [e.g. CASIMIR (Coordination and Sustainability of Mouse Informatics Resources)3 and ENFIN4] have identified this problem and are producing ‘database of databases’ (registries) for their field of expertise.5
A registry of resources needs to be more than just a list of databases and textual descriptions to be useful to the biological and bioinformatics communities. To achieve its aim of helping scientists find the most relevant resource for their needs, it needs to provide at the very least browsing and searching by the type of data contained in each resource, i.e. the biological scope of the resource. A typical approach, as used by all the registries described above and the MRB (Mouse Resource Browser)6 registry developed by a number of the authors of this article, is for a community to define a list of categories (a controlled vocabulary) that covers their scientific domain and then to tag each resource with one or more of these terms. Use of existing and newly developed ontologies for these tags would certainly facilitate future interoperability of the various registries being developed.
While developing MRB, user feedback suggested that it would be helpful if users could go beyond simple categorization of the scope of resources to discover metadata describing database operations and functionality. We therefore set out to capture the utility, accessibility and ease of use of a resource, along with its potential interoperability with other tools and databases. The types of questions that we wanted to be able to answer from this metadata included whether the resource uses automated or manual curation, how often it updates and whether there is a way to track back to different versions, does it provide good technical documentation and user support, does it use recognized standards to record and structure its data, and finally does it go beyond simple web browsing to allow programmatic access and output in standard formats?
Data are always easier to capture and search if a consistent standard is used and we therefore developed a Database Description Framework (DDF; Table 1) as part of the CASIMIR project. Although produced for the MRB, the DDF is generically applicable to any biological database and can be adapted for the requirements of any biological community. For each heading or category, there is a three-tier assessment criterion, a number chosen for simplicity and ease of use. The aim of the DDF is not to make ‘value judgements’ about a resource, but to summarise what it does and what functionalities it supports, with the categories simply reflecting the degree of complexity or sophistication of the database. What is useful or relevant for some databases need not be so for others, and each needs to be assessed in terms of its own remit and user community. The DDF is also intended to be helpful in disseminating and supporting good database practice, in providing backing for resources aspiring to improve the levels of their service, and in giving objective criteria that can be used by external assessors to measure a resource’s progress towards their stated goals.
The CASIMIR Database Description Framework (DDF)
| Category . | Level 1 . | Level 2 . | Level 3 . |
|---|---|---|---|
| Quality and Consistency | No explicit process for assuring consistency | Process for assuring consistency, automatic curation only | Process for assuring consistency with manual curation |
| Currency | Closed legacy database or last update more than a year ago | Updates or versions more than once a year | Updates or versions more than once a month |
| Accessibility | Access via browser only | Access via browser and database reports or database dumps | Access via browser and programmatic access (well defined API, SQL access or web services) |
| Output formats | HTML or similar to browser only | HTML or similar to browser and sparse standard file formats, e.g. FASTA | HTML or similar to browser and rich standard file formats, e.g. XML, SBML (Systems Biology Markup Language) |
| Technical documentation | Written text only | Written text and formal structured description, e.g. automatically generated API docs (JavaDoc), DDL (Data Description Language), DTD (Document Type Definition), UML (Unified Modelling Language), etc. | Written text and formal structured description and tutorials or demonstrations on how to use them |
| Data representation standards | Data coded by local formalism only | Some data coded by a recognised controlled vocabulary, ontology or use of minimal information standards (MIBBI) | General use of both recognised vocabularies or ontologies, and minimal information standards (MIBBI) |
| Data structure standards | Data structured with local model only | Data structured with formal model, e.g. an XML schema | Use of recognised standard model, e.g. FUGE |
| User support | User documentation only | User documentation and Email/web form help desk function | User documentation as well as a personal contact help desk function/training |
| Versioning | No provision | Previous version of database available but no tracking of entities between versions | Previous version of database available and tracking of entities between versions |
| Category . | Level 1 . | Level 2 . | Level 3 . |
|---|---|---|---|
| Quality and Consistency | No explicit process for assuring consistency | Process for assuring consistency, automatic curation only | Process for assuring consistency with manual curation |
| Currency | Closed legacy database or last update more than a year ago | Updates or versions more than once a year | Updates or versions more than once a month |
| Accessibility | Access via browser only | Access via browser and database reports or database dumps | Access via browser and programmatic access (well defined API, SQL access or web services) |
| Output formats | HTML or similar to browser only | HTML or similar to browser and sparse standard file formats, e.g. FASTA | HTML or similar to browser and rich standard file formats, e.g. XML, SBML (Systems Biology Markup Language) |
| Technical documentation | Written text only | Written text and formal structured description, e.g. automatically generated API docs (JavaDoc), DDL (Data Description Language), DTD (Document Type Definition), UML (Unified Modelling Language), etc. | Written text and formal structured description and tutorials or demonstrations on how to use them |
| Data representation standards | Data coded by local formalism only | Some data coded by a recognised controlled vocabulary, ontology or use of minimal information standards (MIBBI) | General use of both recognised vocabularies or ontologies, and minimal information standards (MIBBI) |
| Data structure standards | Data structured with local model only | Data structured with formal model, e.g. an XML schema | Use of recognised standard model, e.g. FUGE |
| User support | User documentation only | User documentation and Email/web form help desk function | User documentation as well as a personal contact help desk function/training |
| Versioning | No provision | Previous version of database available but no tracking of entities between versions | Previous version of database available and tracking of entities between versions |
The CASIMIR Database Description Framework (DDF)
| Category . | Level 1 . | Level 2 . | Level 3 . |
|---|---|---|---|
| Quality and Consistency | No explicit process for assuring consistency | Process for assuring consistency, automatic curation only | Process for assuring consistency with manual curation |
| Currency | Closed legacy database or last update more than a year ago | Updates or versions more than once a year | Updates or versions more than once a month |
| Accessibility | Access via browser only | Access via browser and database reports or database dumps | Access via browser and programmatic access (well defined API, SQL access or web services) |
| Output formats | HTML or similar to browser only | HTML or similar to browser and sparse standard file formats, e.g. FASTA | HTML or similar to browser and rich standard file formats, e.g. XML, SBML (Systems Biology Markup Language) |
| Technical documentation | Written text only | Written text and formal structured description, e.g. automatically generated API docs (JavaDoc), DDL (Data Description Language), DTD (Document Type Definition), UML (Unified Modelling Language), etc. | Written text and formal structured description and tutorials or demonstrations on how to use them |
| Data representation standards | Data coded by local formalism only | Some data coded by a recognised controlled vocabulary, ontology or use of minimal information standards (MIBBI) | General use of both recognised vocabularies or ontologies, and minimal information standards (MIBBI) |
| Data structure standards | Data structured with local model only | Data structured with formal model, e.g. an XML schema | Use of recognised standard model, e.g. FUGE |
| User support | User documentation only | User documentation and Email/web form help desk function | User documentation as well as a personal contact help desk function/training |
| Versioning | No provision | Previous version of database available but no tracking of entities between versions | Previous version of database available and tracking of entities between versions |
| Category . | Level 1 . | Level 2 . | Level 3 . |
|---|---|---|---|
| Quality and Consistency | No explicit process for assuring consistency | Process for assuring consistency, automatic curation only | Process for assuring consistency with manual curation |
| Currency | Closed legacy database or last update more than a year ago | Updates or versions more than once a year | Updates or versions more than once a month |
| Accessibility | Access via browser only | Access via browser and database reports or database dumps | Access via browser and programmatic access (well defined API, SQL access or web services) |
| Output formats | HTML or similar to browser only | HTML or similar to browser and sparse standard file formats, e.g. FASTA | HTML or similar to browser and rich standard file formats, e.g. XML, SBML (Systems Biology Markup Language) |
| Technical documentation | Written text only | Written text and formal structured description, e.g. automatically generated API docs (JavaDoc), DDL (Data Description Language), DTD (Document Type Definition), UML (Unified Modelling Language), etc. | Written text and formal structured description and tutorials or demonstrations on how to use them |
| Data representation standards | Data coded by local formalism only | Some data coded by a recognised controlled vocabulary, ontology or use of minimal information standards (MIBBI) | General use of both recognised vocabularies or ontologies, and minimal information standards (MIBBI) |
| Data structure standards | Data structured with local model only | Data structured with formal model, e.g. an XML schema | Use of recognised standard model, e.g. FUGE |
| User support | User documentation only | User documentation and Email/web form help desk function | User documentation as well as a personal contact help desk function/training |
| Versioning | No provision | Previous version of database available but no tracking of entities between versions | Previous version of database available and tracking of entities between versions |
caBIG, the NCI Cancer Bioinformatics grid7 has produced a similar framework for capturing resource metadata but with a stronger focus on the technical assessment of the resources that wish to participate in the project. As caBIG has a well-defined set of tasks and a user community tied to the specific vision and funding, their categories and levels are less generic than those in the DDF and more focused on assessing whether databases reach a required level of interoperability to interact with the other components of this particular project.
Registries of web services
As well as capturing the scope and database practices of resources, registries need to be explicit about the modes of programmatic access that databases provide (e.g. web services) as these are increasingly used to build database networks and cyberinfrastructure.8–10 This technical information is often hard to find in publications or even on database web sites, but can radically change the strategy adopted by bioinformaticians needing to access the database—for example, integration into automated or semi-automated work flows using Taverna11 such as that developed by CASIMIR.12 Unfortunately, traditional web-service description languages such as WSDL do not provide the required detail on the biological context of the inputs and outputs of each service to allow automated data and service integration. Biocatalogue13 and its predecessor, the EMBRACE service registry14, address this lack of semantics by providing sites for the registration, curation, discovery and monitoring of web services for the whole biological community. Curation of information about web services is open to anyone and uses a combination of free text, tags, ontology terms and example values to describe what each service does, the type of web service (REST, SOAP, soaplab) and in particular the input and outputs in terms of what type of biological data and data formats are expected. Biocatalogue clearly addresses a vital requirement of the community and already some 1173 services have been annotated, despite the project only running for just over a year. Having a single, well-designed solution rather than multiple competing efforts is likely to improve further uptake, and we propose that all registries of databases utilize Biocatalogue to annotate the services provided by their resources rather than separately performing this task.
Dissemination issues and solutions
Capturing metadata as described for the DDF or the Biocatalogue project is not easy. Our initial DDF metadata for over 220 resources was captured as part of a detailed MRB questionnaire sent to each resource, and active manual curation had to be used to fill in the gaps in responses. This is expensive and time-consuming and, after the first pass, there is a requirement to keep the captured data up to date, and this is not easily met.
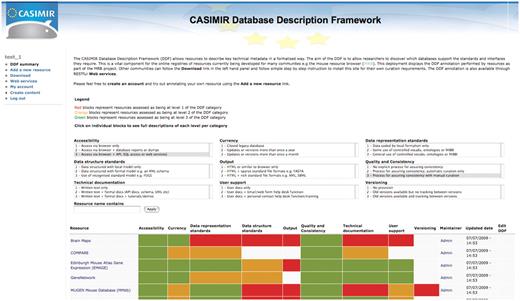
To eliminate the cost of a central curation effort, it would be much better if each resource curated their own metadata and made it accessible to the wider scientific community. As an example of this, we produced a DDF extension to the Drupal content management system (http://drupal.org), which allows curators to log-in and categorize their databases in terms of DDF categories and levels using a simple web form. The resulting metadata is then browsable and searchable either through a web interface or programmatically through RESTful web services. An example deployment is viewable at www.casimir.org.uk/casimir_ddf (Figure 1) and is currently populated with the metadata for the MRB project. We encourage interested readers to visit our site and for maintainers of resources to curate their metadata using it. The Drupal framework is easily extensible to allow curation of other data associated with each resource, so allowing the production of a customisable community registry. The system is expected to be of great value to communities developing registry resources or individual informaticians wanting to establish quickly which features a database provides (the software is freely available under an open-source license). The REST web services allow a central DDF portal to be established offering the collection and sharing of data from individual database registries as well as avoiding redundancy in curation efforts.
The DDF query and annotation tool. This tool allows any user to browse a set of resources that have been annotated using the DDF categories. Searches for resources by DDF category and level are also possible. In addition, resource maintainers can log-in and edit their existing annotations or annotate a new resource using a simple web form. This tool is freely available and easy to install for other communities that wish to create their own registry of resources.
Biocatalogue have used a combination of central and community curation from the outset to capture data on web services and the large number of services already described is testament to such an approach. Again, the provision of easy to use web tools that suggest particular tags and ontology terms to use in the annotation increases the likelihood of achieving a high level of community engagement and annotation quality.
Community curation requires pro-active participation. Communities need to acknowledge; (i) a central site where they can find relevant resources would be useful, and; (ii) the only practical means of achieving this is for each database to self-curate its entry using a clearly articulated and standardized set of benchmarks and tools such as provided by the DDF and Biocatalogue solutions. Individual resources would also benefit from this small amount of curation effort as the central registry will direct users to them, who might not previously have known about their resource. Although the creators and maintainers of a resource are best placed to describe the associated metadata, a self-curation approach can raise data quality issues, but these should be minimized if the annotation tools are well designed i.e. fast and easy to use, with clear descriptions of what is being asked for, and responses presented as a lists of terms rather than free text. However, even with a well-designed annotation tool, registries are still likely to require some central curation for validating submitted data (e.g. the DDF tool allows administrator level access to check new submissions).
In summary, there is now a clear need for registries to be built that address biological categorization of databases and services, annotate any services provided and capture metadata on database best practises. Considerable progress has been made on standardizing the capture of each of these by such approaches as the DDF and Biocatalogue, but the community would benefit from coordination to produce full registries combining all these approaches. However, the value of a standard is dependent on its uptake by the community as can be seen, for example, in the MIBBI family of minimal information standards.15 Uptake of a standard is, of course, as much a social issue as one of producing the right technologies for the community. Here, support from funding agencies and journals will be vital in establishing the practice of publishing database and services metadata. All curators can enhance the value of their databases by posting a minimal amount of information about their resource on a community site. The task has minimal cost, but will provide considerable value to investigators, database developers, informaticians and funding agencies.
Funding
The work described above was supported by Seventh Framework Programme of the European Commission contracts to CASIMIR (LSHG-CT-2006-037811) and ENFIN (LSHG-CT-2005-518254). Funding for open access charge: CASIMIR LSHG-CT-2006-037811.
Conflict of interest. None declared.
References
Author notes
Reference 13 in an earlier version of this paper was incorrect. The author apologizes for this error.