-
PDF
- Split View
-
Views
-
Cite
Cite
Philip Jones, David Binns, Conor McMenamin, Craig McAnulla, Sarah Hunter, The InterPro BioMart: federated query and web service access to the InterPro Resource, Database, Volume 2011, 2011, bar033, https://doi.org/10.1093/database/bar033
Close - Share Icon Share
Abstract
The InterPro BioMart provides users with query-optimized access to predictions of family classification, protein domains and functional sites, based on a broad spectrum of integrated computational models (‘signatures’) that are generated by the InterPro member databases: Gene3D, HAMAP, PANTHER, Pfam, PIRSF, PRINTS, ProDom, PROSITE, SMART, SUPERFAMILY and TIGRFAMs. These predictions are provided for all protein sequences from both the UniProt Knowledge Base and the UniParc protein sequence archive. The InterPro BioMart is supplementary to the primary InterPro web interface (http://www.ebi.ac.uk/interpro), providing a web service and the ability to build complex, custom queries that can efficiently return thousands of rows of data in a variety of formats. This article describes the information available from the InterPro BioMart and illustrates its utility with examples of how to build queries that return useful biological information.
Database URL: http://www.ebi.ac.uk/interpro/biomart/martview.
Introduction
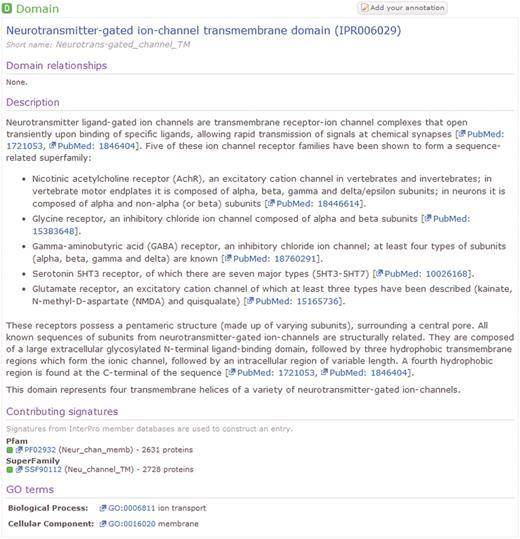
The InterPro Resource (http://www.ebi.ac.uk/interpro) (1) provides an integrated set of computational models (or signatures) for protein family classification and the prediction of structural and functional domains, sites and repeats. The predictive signatures are built by the 11 InterPro member databases that, together with the InterPro team at the EBI, comprise the InterPro Consortium. The member databases are Gene3D (2), HAMAP (3), PANTHER (4), Pfam (5), PIRSF (6), PRINTS (7), ProDom (8), PROSITE (9), SMART (10), SUPERFAMILY (11) and TIGRFAMs (12). The InterPro team at the EBI integrates the predictive signatures from these member databases into ‘InterPro Entries’. Each entry may include one or more signatures that either identify the same feature, or classify proteins into the same family. Additionally, entries are collated into two biologically principled hierarchies, one of which describes protein families, the other protein domains. InterPro entries are curated by a team of experts in a variety of fields in Biology. The curation process includes the creation of entries, the structuring of the entry hierarchies, the provision of detailed abstracts describing each entry and the addition of useful cross-references to other databases and ontologies. An example InterPro Entry, as viewed on the main InterPro website, is illustrated in Figure 1. This example entry comprises two member database signatures, one from Pfam and the other from SuperFamily. In total, this InterPro entry matches 2753 UniProtKB protein sequences.
An example human-curated InterPro entry, illustrating the detailed description provided for the entry and cross references to the GO and the member database signatures from which the entry is composed.
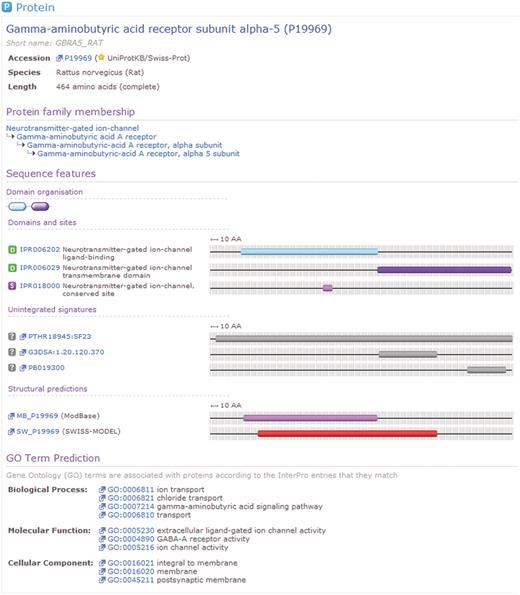
The integration described above is useful because the individual member databases have distinct but overlapping interests and use a number of different algorithms and modeling techniques. From the perspective of the biologist or bioinformatician wishing to use these predictive techniques, InterPro allows consideration of all of the available signatures from a single resource, without the need to be concerned with differences or overlap between the foci of the individual member databases. As well as integrating the member database signatures, InterPro calculates matches to these signatures for the whole of the UniProt Knowledge Base (UniProtKB, http://www.uniprot.org) and the UniParc sequence archive (13). Figure 2 illustrates a set of matches to a single UniProtKB protein sequence, which matches three InterPro entries. The matches of InterPro signatures and entries to the sequences in UniProtKB are available from the main InterPro website as well as from the InterPro BioMart, however, at the time of writing, the matches to UniParc sequences are only available from the BioMart. It is expected that UniParc matches will be included in a future version of the main InterPro website. The InterPro BioMart is built on the technology developed by the BioMart project (http://www.biomart.org) (14, 15), a collaboration between the Ontario Institute for Cancer Research (OICR) and the European Bioinformatics Institute (EBI). The InterPro BioMart is available at http://www.ebi.ac.uk/interpro/biomart/martview. It is also incorporated into the BioMart Central Server at http://www.biomart.org/biomart/martview (16).
A protein for which matches have been calculated by InterPro. For this sequence, InterPro provides a prediction of protein family membership, an overview of the domain organization and the details of matches to member database signatures. At the foot of the view can be seen associated GO terms, based upon the calculated matches to InterPro entries.
The adoption of BioMart as a mechanism to share the data in InterPro has been motivated by the benefits that BioMart brings: the ability to build complex filters on the data; the facility to select specifically which data types are returned (equivalent to the columns of a spreadsheet); the capacity of BioMart to handle queries that return many thousands of rows of data and the provision of a web service with an associated data federation mechanism.
Data content
The InterPro BioMart provides three data sources: ‘InterPro Entry Annotation’, ‘UniProtKB Protein Matches’ and ‘UniParc Protein Matches’.
Match information can be obtained from both the ‘InterPro Entry Annotation’ data source and the ‘UniProtKB Protein Matches’ data source. These two data sources provide a different slant on the contents of InterPro, as described below.
The ‘InterPro Entry Annotation’ data source focuses on descriptions of the InterPro entries and the hierarchical relationships between them. The user can therefore build filters using this annotation and retrieve more detailed information, such as assigned Gene Ontology (19) terms and cross-references to other, related databases. The ‘Query Examples’ section below illustrates a potential application of this data set.
The ‘UniProtKB Protein Matches’ data set is focused on the UniProtKB protein entity, allowing queries to be built based on attributes of the protein sequence, including options to filter on the taxonomic group annotated on the sequence. This data set also provides the opportunity to retrieve match information with respect to member database signatures as well as summarized match information, described as ‘supermatches’ in the BioMart. A ‘supermatch’ is determined where one or more member database signatures that have been integrated together into the same entry have overlapping matches to the protein in the same region of the sequence. The start and stop coordinates of the InterPro entry ‘supermatch’ are then calculated as the most extreme bounds of the matches of all the member databases’ signatures comprising the entry.
Finally the ‘UniParc Protein Matches’ data set provides equivalent information to the ‘UniProtKB Protein matches’ data set, coordinated on sequences included in the UniParc database, a non-redundant, historical archive of protein sequences extracted from public databases. At the time of writing, the UniParc database includes 25.6 million unique sequences; the InterPro match calculation pipeline is run against all of these sequences and the results are made available from this BioMart data set. This service allows matches to be returned for sequences that are present in (for example) model organism protein sequence databases which are not yet represented in UniProtKB. For users interested in matches for specific protein sequences, this data set supports filtering by UniParc ID or sequence checksum (CRC-64 or MD5), as does the ‘UniProtKB Protein Matches’ data set. If the user wishes to query using protein accessions or identifiers from third-party sequence databases, various services are available that allow protein identifier cross-referencing, including the Protein Identifier Cross Reference service, PICR (http://www.ebi.ac.uk/Tools/picr/) (20) and the UniProt ID mapping service (http://www.uniprot.org/). Both of these services can be used to convert protein identifiers or accessions from a large number of protein sequence databases to UniParc sequence identifiers.
The three InterPro BioMart data sources include matches to the full taxonomy range in UniProtKB or UniParc. In this respect, the InterPro BioMart is different in structure to the Ensembl BioMart (http://www.ensembl.org/biomart/martview) (17, 18) which is organized into species-specific data sets.
Services supported by the InterPro BioMart
The InterPro BioMart is used to extend the functionality of the primary InterPro web interface, providing BioMart ‘canned queries’ for InterPro entries and for matched proteins. This allows data to be downloaded in tab- or comma-separated values format, suitable for computational analysis.
The InterPro BioMart web service is the data source behind the InterPro Distributed Annotation System (DAS) service (21), available from http://www.ebi.ac.uk/das-srv/interpro/das. This DAS service provides four DAS sources that query the BioMart.
‘InterPro’, which contains all InterPro member database signature matches to UniProtKB protein sequences.
‘InterPro-matches-overview’ that provides the maximum extent of the matches from all signatures that are integrated into a single InterPro entry against UniProtKB protein sequences. These are the ‘supermatch’ matches described in the BioMart ‘UniProtKB Protein Matches’ data set.
‘InterPro-UniParc-matches’ that provides match information for protein sequences identified using UniParc identifiers.
‘InterPro-S4’ is used to provide protein family classification to the new EBI Search Service and is therefore part of the wider programme of data integration at the EBI.
Query examples
In common with all BioMart implementations, the InterPro BioMart enables the construction of simple queries as well as complex, multi-faceted queries where the data is filtered on several criteria. Where multiple filters are applied, records are returned that meet all of the filter criteria (i.e. ‘AND’ logic is applied across the filters). The user is able to specify precisely which data attributes should be returned, equivalent to columns in a spreadsheet.
Users should be aware that the structure of a BioMart database, which is highly redundant to facilitate high query speed, can result in redundancy in the results reported. The presence of repeated rows of results in the output depends on the construction of the query and the structure of the underlying BioMart tables. The circumstances under which this may occur are not self-evident. The authors therefore recommend the use of the ‘Unique results only’ option when querying the BioMart, which removes repeated rows of results.
To demonstrate the utility of the InterPro BioMart, here we present several biologically relevant queries
Query #1. ‘Which Pfam signatures does InterPro integrate into “family” entries?’
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | InterPro Entry Type: ‘Family’ | InterPro Entry Accession |
| Source Signature Database: ‘Pfam’ | InterPro Entry Short Name | |
| Signature Accession | ||
| Signature ID (Name) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | InterPro Entry Type: ‘Family’ | InterPro Entry Accession |
| Source Signature Database: ‘Pfam’ | InterPro Entry Short Name | |
| Signature Accession | ||
| Signature ID (Name) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | InterPro Entry Type: ‘Family’ | InterPro Entry Accession |
| Source Signature Database: ‘Pfam’ | InterPro Entry Short Name | |
| Signature Accession | ||
| Signature ID (Name) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | InterPro Entry Type: ‘Family’ | InterPro Entry Accession |
| Source Signature Database: ‘Pfam’ | InterPro Entry Short Name | |
| Signature Accession | ||
| Signature ID (Name) |
The Pfam database contains a broad spectrum of hidden Markov models that can be used to predict both family classification and domain organization. The InterPro curation team has integrated >96% of Pfam signatures into InterPro at the time of writing. During integration, InterPro assigns a ‘type’ to an InterPro entry and by extension, its signatures, dependent on what is being represented (a Family, Domain, Site or Repeat). Using the BioMart, it is possible to return the full set of integrated Pfam signatures that InterPro considers to be of type ‘family’. This query can be easily modified to request signatures built by any of the member databases that fit into any of the available InterPro entry types. Entry type filters include ‘Active_site’, ‘Binding_site’, ‘Conserved_site’, ‘Domain’, ‘Family’, ‘PTM’ (Post Translational Modification) and ‘Repeat’. Each InterPro entry has exactly one type and consequently all integrated member database signatures also have one type, as assigned by the InterPro curation team.
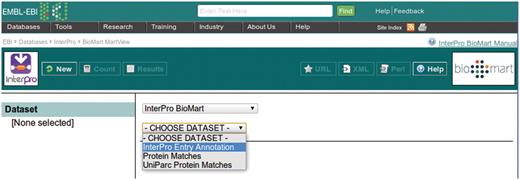
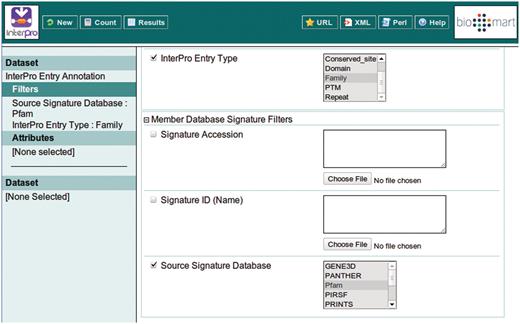
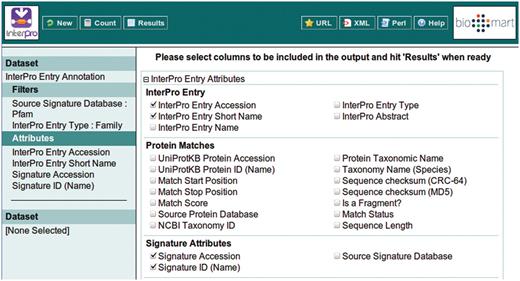
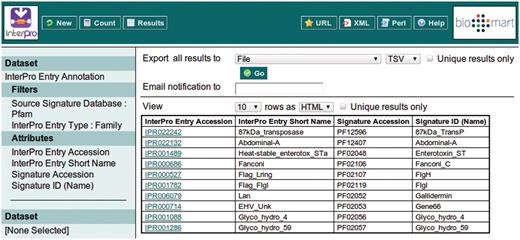
This example query is illustrated with a series of screen shots. Figure 3 illustrates selection of the InterPro Entry Annotation data set. Following selection of this data set, the user is able to select filters and attributes (in whichever order they choose). Figure 4 illustrates the selection of the two filters applied in this query, which will restrict the rows of data returned. Figure 5 illustrates the selection of attributes, which are equivalent to the columns of a spreadsheet. Finally, Figure 6 illustrates the results that are obtained when the ‘Results’ button is pressed. Initially, the user is presented with the first 10 matching rows of data, giving an opportunity to refine the query prior to requesting the full set of results.
Building a filter with two components: include results for ‘Family’ entry types that comprise signatures from Pfam.
Selecting the attributes to be included in the BioMart output (equivalent to the columns of a spreadsheet). The ordering of the columns is determined by the order in which the attributes are selected.
Clicking the ‘Results’ button at the top of the interface provides the first 10 results matching the query, to allow the query to be modified or improved.
Query #2. ‘Which GO terms are mapped to PROSITE signatures in InterPro (i.e. Can I retrieve a PROSITE2GO mapping?)’
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | Source Signature Database : ‘PROSITE patterns’ and ‘PROSITE Profiles’ (CTRL click to select both). | InterPro Entry Accession |
| Signature Accession | ||
| GO ID | ||
| GO Term Name | ||
| GO Root Term (Process/Component/Function) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | Source Signature Database : ‘PROSITE patterns’ and ‘PROSITE Profiles’ (CTRL click to select both). | InterPro Entry Accession |
| Signature Accession | ||
| GO ID | ||
| GO Term Name | ||
| GO Root Term (Process/Component/Function) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | Source Signature Database : ‘PROSITE patterns’ and ‘PROSITE Profiles’ (CTRL click to select both). | InterPro Entry Accession |
| Signature Accession | ||
| GO ID | ||
| GO Term Name | ||
| GO Root Term (Process/Component/Function) |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| InterPro Entry Annotation | Source Signature Database : ‘PROSITE patterns’ and ‘PROSITE Profiles’ (CTRL click to select both). | InterPro Entry Accession |
| Signature Accession | ||
| GO ID | ||
| GO Term Name | ||
| GO Root Term (Process/Component/Function) |
A major use of InterPro is the association of GO terms to proteins via the signatures that they match. InterPro provides the ‘InterPro2GO’ mappings as a file that can be downloaded from the FTP site; however, it is difficult to extract subsets of information from this file. In the past, a frequent request from users was the provision of GO term mapping information for a particular member database. With the advent of the BioMart, it is now very easy to provide this information as illustrated above.
Query #3. ‘Which metabolic pathways are associated with proteins matching the InterPro family “Chemokine receptor type 4” (CXCR4, IPR001277)?’
| Data sets . | Filters . | Attributes . |
|---|---|---|
| pathway | Pathway stable ID | |
| Pathway name | ||
| InterPro Entry Annotation | InterPro Entry ID = ‘IPR001277’ | InterPro Entry Accession |
| InterPro Entry Name | ||
| UniProtKB Protein Accession | ||
| UniProtKB Protein ID (Name) | ||
| Source Signature Database | ||
| Signature Accession | ||
| Signature ID (Name) | ||
| Match Start Position | ||
| Match Stop Position |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| pathway | Pathway stable ID | |
| Pathway name | ||
| InterPro Entry Annotation | InterPro Entry ID = ‘IPR001277’ | InterPro Entry Accession |
| InterPro Entry Name | ||
| UniProtKB Protein Accession | ||
| UniProtKB Protein ID (Name) | ||
| Source Signature Database | ||
| Signature Accession | ||
| Signature ID (Name) | ||
| Match Start Position | ||
| Match Stop Position |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| pathway | Pathway stable ID | |
| Pathway name | ||
| InterPro Entry Annotation | InterPro Entry ID = ‘IPR001277’ | InterPro Entry Accession |
| InterPro Entry Name | ||
| UniProtKB Protein Accession | ||
| UniProtKB Protein ID (Name) | ||
| Source Signature Database | ||
| Signature Accession | ||
| Signature ID (Name) | ||
| Match Start Position | ||
| Match Stop Position |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| pathway | Pathway stable ID | |
| Pathway name | ||
| InterPro Entry Annotation | InterPro Entry ID = ‘IPR001277’ | InterPro Entry Accession |
| InterPro Entry Name | ||
| UniProtKB Protein Accession | ||
| UniProtKB Protein ID (Name) | ||
| Source Signature Database | ||
| Signature Accession | ||
| Signature ID (Name) | ||
| Match Start Position | ||
| Match Stop Position |
The InterPro BioMart is federated with the Reactome BioMart (22, 23) from which the ‘pathway’ data set derives. Reactome describes ‘reactions, pathways and biological processes’ and as such, can provide valuable biological insight if married to the data in InterPro.
Query #4. ‘In which tissues have proteins matching the InterPro family “Neural cell adhesion” (IPR009138) been identified by mass spectrometry?’
| Data sets . | Filters . | Attributes . |
|---|---|---|
| PRIDE | PRIDE Experiment Accession | |
| Experiment Title | ||
| Sample Name | ||
| Taxonomy Term (NEWT/NCBI Taxon) | ||
| Taxonomy ID (NEWT/NCBI Taxon) | ||
| Tissue Ontology Term (BRENDA) | ||
| BRENDA ID (Tissue) | ||
| Cell Type Term (CL) | ||
| CL ID (Cell Type) | ||
| Gene Ontology Term (GO) | ||
| GO ID (Gene Ontology) | ||
| InterPro Entries | InterPro Entry ID = ‘IPR009138’ | InterPro Entry Accession |
| InterPro Entry Name |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| PRIDE | PRIDE Experiment Accession | |
| Experiment Title | ||
| Sample Name | ||
| Taxonomy Term (NEWT/NCBI Taxon) | ||
| Taxonomy ID (NEWT/NCBI Taxon) | ||
| Tissue Ontology Term (BRENDA) | ||
| BRENDA ID (Tissue) | ||
| Cell Type Term (CL) | ||
| CL ID (Cell Type) | ||
| Gene Ontology Term (GO) | ||
| GO ID (Gene Ontology) | ||
| InterPro Entries | InterPro Entry ID = ‘IPR009138’ | InterPro Entry Accession |
| InterPro Entry Name |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| PRIDE | PRIDE Experiment Accession | |
| Experiment Title | ||
| Sample Name | ||
| Taxonomy Term (NEWT/NCBI Taxon) | ||
| Taxonomy ID (NEWT/NCBI Taxon) | ||
| Tissue Ontology Term (BRENDA) | ||
| BRENDA ID (Tissue) | ||
| Cell Type Term (CL) | ||
| CL ID (Cell Type) | ||
| Gene Ontology Term (GO) | ||
| GO ID (Gene Ontology) | ||
| InterPro Entries | InterPro Entry ID = ‘IPR009138’ | InterPro Entry Accession |
| InterPro Entry Name |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| PRIDE | PRIDE Experiment Accession | |
| Experiment Title | ||
| Sample Name | ||
| Taxonomy Term (NEWT/NCBI Taxon) | ||
| Taxonomy ID (NEWT/NCBI Taxon) | ||
| Tissue Ontology Term (BRENDA) | ||
| BRENDA ID (Tissue) | ||
| Cell Type Term (CL) | ||
| CL ID (Cell Type) | ||
| Gene Ontology Term (GO) | ||
| GO ID (Gene Ontology) | ||
| InterPro Entries | InterPro Entry ID = ‘IPR009138’ | InterPro Entry Accession |
| InterPro Entry Name |
The InterPro BioMart is also federated with the PRIDE BioMart. PRIDE is the ‘Proteomics Identifications Database’, which contains identifications of proteins and peptides arising from mass spectrometry. The two BioMarts are linked via UniProtKB protein accessions, so this query returns information about identifications of the proteins that match the member database signatures integrated into InterPro Entry IPR009138.
Discussion and future directions
The InterPro BioMart has proven a valuable addition to the InterPro software infrastructure, supporting new tools, such as the InterPro DAS service, as well as providing an efficient route to answer queries from the InterPro user community. The BioMart has furnished InterPro with a web service, for which robust APIs exist in several languages (including both Perl and Java).
Additionally, BioMart provides a substantial resource for bioinformaticians to query InterPro, alongside the federated databases UniProtKB, Reactome and PRIDE. (See Table 1, which describes these Bioinformatics resources).
External data sources included in the InterPro BioMart
| Source . | URL . | BioMart URL . | Description of contents . |
|---|---|---|---|
| UniProtKB | http://www.uniprot.org | http://www.ebi.ac.uk/ uniprot/biomart/martview | A comprehensive, high quality and freely accessible resource of protein sequence and functional information, comprising the human-curated Swiss-Prot data set and the automatically annotated TrEMBL data set. |
| PRIDE | http://www.ebi.ac.uk/pride | http://www.ebi.ac.uk/pride/biomart/martview | A database of identifications of proteins and peptides, arising from mass spectrometry-based proteomics. |
| Reactome Pathway Database | http://www.reactome.org | http://www.reactome.org/cgi-bin/mart | A human-curated database of biological pathways, focusing on human pathways, but providing automated prediction of pathways in other species. |
| Source . | URL . | BioMart URL . | Description of contents . |
|---|---|---|---|
| UniProtKB | http://www.uniprot.org | http://www.ebi.ac.uk/ uniprot/biomart/martview | A comprehensive, high quality and freely accessible resource of protein sequence and functional information, comprising the human-curated Swiss-Prot data set and the automatically annotated TrEMBL data set. |
| PRIDE | http://www.ebi.ac.uk/pride | http://www.ebi.ac.uk/pride/biomart/martview | A database of identifications of proteins and peptides, arising from mass spectrometry-based proteomics. |
| Reactome Pathway Database | http://www.reactome.org | http://www.reactome.org/cgi-bin/mart | A human-curated database of biological pathways, focusing on human pathways, but providing automated prediction of pathways in other species. |
External data sources included in the InterPro BioMart
| Source . | URL . | BioMart URL . | Description of contents . |
|---|---|---|---|
| UniProtKB | http://www.uniprot.org | http://www.ebi.ac.uk/ uniprot/biomart/martview | A comprehensive, high quality and freely accessible resource of protein sequence and functional information, comprising the human-curated Swiss-Prot data set and the automatically annotated TrEMBL data set. |
| PRIDE | http://www.ebi.ac.uk/pride | http://www.ebi.ac.uk/pride/biomart/martview | A database of identifications of proteins and peptides, arising from mass spectrometry-based proteomics. |
| Reactome Pathway Database | http://www.reactome.org | http://www.reactome.org/cgi-bin/mart | A human-curated database of biological pathways, focusing on human pathways, but providing automated prediction of pathways in other species. |
| Source . | URL . | BioMart URL . | Description of contents . |
|---|---|---|---|
| UniProtKB | http://www.uniprot.org | http://www.ebi.ac.uk/ uniprot/biomart/martview | A comprehensive, high quality and freely accessible resource of protein sequence and functional information, comprising the human-curated Swiss-Prot data set and the automatically annotated TrEMBL data set. |
| PRIDE | http://www.ebi.ac.uk/pride | http://www.ebi.ac.uk/pride/biomart/martview | A database of identifications of proteins and peptides, arising from mass spectrometry-based proteomics. |
| Reactome Pathway Database | http://www.reactome.org | http://www.reactome.org/cgi-bin/mart | A human-curated database of biological pathways, focusing on human pathways, but providing automated prediction of pathways in other species. |
It is intended to federate the InterPro BioMart with the new UniParc BioMart that is under development at the EBI. This will allow the InterPro BioMart to be queried using identifiers and accessions from a large variety of protein sequence databases other than the UniProtKB, including several model organism databases.
Funding
Biotechnology and Biological Sciences Research Council's Bioinformatics and Biological Resources Fund (grant number BB/F010508/1); European Union under the program ‘FP7 capacities: Scientific Data Repositories’; The working title for the project is IMproving Protein Annotation and Co-ordination using Technology (IMPACT) (grant number 213037). Funding for open access charge: European Union under the program ‘FP7 capacities: Scientific Data Repositories’; The working title for the project is IMproving Protein Annotation and Co-ordination using Technology (IMPACT) (grant number 213037).
Conflict of interest. None declared.
Acknowledgements
The authors would particularly like to acknowledge the continuing support of the InterPro Consortium member databases and the support of the BioMart development team who have given invaluable guidance and assistance with constructing the InterPro BioMart.