-
PDF
- Split View
-
Views
-
Cite
Cite
Rama Balakrishnan, Julie Park, Kalpana Karra, Benjamin C. Hitz, Gail Binkley, Eurie L. Hong, Julie Sullivan, Gos Micklem, J. Michael Cherry, YeastMine—an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit, Database, Volume 2012, 2012, bar062, https://doi.org/10.1093/database/bar062
Close - Share Icon Share
Abstract
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) provides high-quality curated genomic, genetic, and molecular information on the genes and their products of the budding yeast Saccharomyces cerevisiae. To accommodate the increasingly complex, diverse needs of researchers for searching and comparing data, SGD has implemented InterMine (http://www.InterMine.org), an open source data warehouse system with a sophisticated querying interface, to create YeastMine (http://yeastmine.yeastgenome.org). YeastMine is a multifaceted search and retrieval environment that provides access to diverse data types. Searches can be initiated with a list of genes, a list of Gene Ontology terms, or lists of many other data types. The results from queries can be combined for further analysis and saved or downloaded in customizable file formats. Queries themselves can be customized by modifying predefined templates or by creating a new template to access a combination of specific data types. YeastMine offers multiple scenarios in which it can be used such as a powerful search interface, a discovery tool, a curation aid and also a complex database presentation format.
Database URL:http://yeastmine.yeastgenome.org
Introduction
Model organisms such as yeast, flies, worms, zebrafish, rat and mice provide powerful experimental systems that allow access to different aspects of biology, and researchers therefore focus their interests on the model that can best answer their experimental questions. Model organism databases (MODs) carefully curate the literature for experimental results, integrate these data with other information, and provide the resulting database to researchers via the web. The ability to connect experimental results from one organism to another has been a major limitation and thus enabling researchers to query across these MODs would provide a powerful method for discovery.
The Saccharomyces Genome Database (SGD) collects and organizes biological information including genes and their products of the budding yeast Saccharomyces cerevisiae. The role of SGD is to provide all publicly available published experimental results in an integrated format to researchers and educators via the Internet. These data historically include gene specific characterizations such as mutant phenotypes, biochemical analysis and chromosomal location. New experimental methods that assay all gene products at once or capture DNA–protein interactions at nucleotide resolution have dramatically increased the amount of data associated with a gene or chromosomal regions. At SGD these results are integrated into a rich data model that is distributed online. The types of information that are collected include functional data annotated with the Gene Ontology (GO) controlled vocabulary system, mutant phenotypes, genetic and physical interactions, biochemical pathways, gene expression and protein and nucleic acid sequences. SGD provides a range of web-based tools to search and retrieve different types of data (1).
To fully uncover relationships between gene characteristics, researchers need the ability to carefully dissect both the depth and the breadth of available data within and across organisms. Traditionally, data retrieval options at SGD and other MODs allow users to download either one type of data across multiple genes (e.g. all GO annotations for all genes) or multiple data types for a single gene. However, obtaining multiple data types for multiple features often required downloading and merging multiple files from download and FTP sites. To provide sophisticated queries of integrated data, and allow SGD to keep up with the constant increase in the number of data types and the increasing resolution of these data we sought to develop a more advanced tool. The desired tool would be fast, expandable, permit effective queries of complex data and allow the result to be explored in useful file formats. While other resources such as UniProt or the BioMart at Ensembl offer tools to query and retrieve core S. cerevisiae data such as identifiers, sequences and GO annotations, SGD aims to provide comprehensive access to all S. cerevisiae datatypes in an integrated fashion. Our goal is to provide a tool that is concordant with the main SGD website both in terms of data content and frequency with which these data are updated.
Here we describe an environment that has been created at SGD that allows rich data to be navigated via complex queries, connects data of different types, enables exploration of features shared between gene products and follows across to information provided by other MODs. To be able to handle the complexity of emerging scientific data, and the needs of the scientific community to be able to query across multiple data types for their analyses, SGD was motivated to provide novel and customizable ways by which to access the data within the database. We chose to implement YeastMine, an integrated data warehouse with precomputed tables based on the InterMine platform. YeastMine is a fast and flexible data retrieval tool that provides custom search and download capabilities of existing data within SGD, and is adaptable enough to be able to add future data types. All of the data types within the main SGD database are provided via YeastMine giving researchers the ability to query all relevant information pertaining to a gene or set of genes in one tool. All data retrieval results are exportable in various configurable and standard file formats. In addition, the tool allows for list creation and query storage between sessions. Here we will discuss the implementation details of YeastMine at SGD, describe the additional functionalities that YeastMine brings to SGD, and outline examples of data retrieval and download.
Creating YeastMine: an implementation of intermine at SGD
Unique software features of YeastMine
The InterMine (ref. 2, http://www.InterMine.org) software provides an easily implemented solution for data integration and web display. Data integration is accomplished by using a data warehouse model that combines and stores a wide range of data types from multiple sources in denormalized precomputed tables in a PostgreSQL database. By using a data warehouse model, retrieving data from the database is very fast, resulting in exceptional web performance. The PostgreSQL database is rebuilt weekly from the main SGD database. Since data types can be added incrementally, new data in the SGD database can be easily integrated into YeastMine. In addition to the core InterMine environment that is used to extract and load data into YeastMine, existing data parsers can be modified and customized Java-based converters can be written. These features further extend the flexibility of YeastMine to provide access to the breadth and depth of literature-based curation and genome-wide analyses available at SGD.
Integration of S. cerevisiae data in YeastMine
Due to their ease of setup, flexibility of data addition, query speed and robustness, as well as the straightforward nature of customizing its software components, we implemented the InterMine platform at SGD to create YeastMine, a tool that integrates the S. cerevisiae data described in Table 1. The range of data types include the genomic and protein sequences for the genes, chromosomal features and genetic loci identified in S. cerevisiae as well as literature, phenotypes, interactions, GO annotations, pathways, homologs and gene expression data that provide insight into a gene product's biological role.
Data types integrated into YeastMine, the source project of the annotations and means by which these data can be accessed from detailed web pages or downloadable files from http://downloads.yeastgenome.org
| Data type . | Source . | Web page at SGD . | Downloadable files (http:// downloads.yeastgenome.org/) . |
|---|---|---|---|
| Basic gene information (description of gene function, gene names) | SGD | Locus Summary | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Chromosomal coordinates, sequence for chromosomal features | SGD | Locus Summary, GBrowse, Gene/Seq resources, PatMatch, BLAST | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Gene ontology annotations | SGD | Locus Summary, GO term | curation/gene_association.sgd |
| Mutant phenotype | SGD | Locus Summary | curation/phenotype_data.tab |
| Interactions | BioGRID | Locus Summary | curation/interaction_data.tab |
| Protein properties | SGD | Locus Protein | curation/protein_properties.tab |
| Biochemical pathways | SGD | Locus Summary, YeastCyc | curation/biochemical_pathways.tab |
| Literature | SGD | Locus Literature Guide, Curated Paper, Textpresso full-text search | curation/gene_literature.tab |
| Gene expression | SPELL | Locus Expression, SPELL | published_datasets/ |
| Homologs | TreeFam | Not currently available in SGD | genomics/homology/ |
| Data type . | Source . | Web page at SGD . | Downloadable files (http:// downloads.yeastgenome.org/) . |
|---|---|---|---|
| Basic gene information (description of gene function, gene names) | SGD | Locus Summary | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Chromosomal coordinates, sequence for chromosomal features | SGD | Locus Summary, GBrowse, Gene/Seq resources, PatMatch, BLAST | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Gene ontology annotations | SGD | Locus Summary, GO term | curation/gene_association.sgd |
| Mutant phenotype | SGD | Locus Summary | curation/phenotype_data.tab |
| Interactions | BioGRID | Locus Summary | curation/interaction_data.tab |
| Protein properties | SGD | Locus Protein | curation/protein_properties.tab |
| Biochemical pathways | SGD | Locus Summary, YeastCyc | curation/biochemical_pathways.tab |
| Literature | SGD | Locus Literature Guide, Curated Paper, Textpresso full-text search | curation/gene_literature.tab |
| Gene expression | SPELL | Locus Expression, SPELL | published_datasets/ |
| Homologs | TreeFam | Not currently available in SGD | genomics/homology/ |
Data types integrated into YeastMine, the source project of the annotations and means by which these data can be accessed from detailed web pages or downloadable files from http://downloads.yeastgenome.org
| Data type . | Source . | Web page at SGD . | Downloadable files (http:// downloads.yeastgenome.org/) . |
|---|---|---|---|
| Basic gene information (description of gene function, gene names) | SGD | Locus Summary | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Chromosomal coordinates, sequence for chromosomal features | SGD | Locus Summary, GBrowse, Gene/Seq resources, PatMatch, BLAST | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Gene ontology annotations | SGD | Locus Summary, GO term | curation/gene_association.sgd |
| Mutant phenotype | SGD | Locus Summary | curation/phenotype_data.tab |
| Interactions | BioGRID | Locus Summary | curation/interaction_data.tab |
| Protein properties | SGD | Locus Protein | curation/protein_properties.tab |
| Biochemical pathways | SGD | Locus Summary, YeastCyc | curation/biochemical_pathways.tab |
| Literature | SGD | Locus Literature Guide, Curated Paper, Textpresso full-text search | curation/gene_literature.tab |
| Gene expression | SPELL | Locus Expression, SPELL | published_datasets/ |
| Homologs | TreeFam | Not currently available in SGD | genomics/homology/ |
| Data type . | Source . | Web page at SGD . | Downloadable files (http:// downloads.yeastgenome.org/) . |
|---|---|---|---|
| Basic gene information (description of gene function, gene names) | SGD | Locus Summary | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Chromosomal coordinates, sequence for chromosomal features | SGD | Locus Summary, GBrowse, Gene/Seq resources, PatMatch, BLAST | curation/SGD_features.tab, curation/saccharomyces_cerevisiae.gff |
| Gene ontology annotations | SGD | Locus Summary, GO term | curation/gene_association.sgd |
| Mutant phenotype | SGD | Locus Summary | curation/phenotype_data.tab |
| Interactions | BioGRID | Locus Summary | curation/interaction_data.tab |
| Protein properties | SGD | Locus Protein | curation/protein_properties.tab |
| Biochemical pathways | SGD | Locus Summary, YeastCyc | curation/biochemical_pathways.tab |
| Literature | SGD | Locus Literature Guide, Curated Paper, Textpresso full-text search | curation/gene_literature.tab |
| Gene expression | SPELL | Locus Expression, SPELL | published_datasets/ |
| Homologs | TreeFam | Not currently available in SGD | genomics/homology/ |
The ability to customize InterMine software allows SGD to incorporate data types that are common to other implementations of InterMine and then extend the data model to provide additional curated details that are uniquely captured at SGD. For example, GO annotations are provided by many database projects and included in their InterMine implementations (2,3). SGD captures the concept of an ‘Annotation Method’ that distinguishes manually curated, high-throughput and computationally predicted GO annotation (4). A parser to load GO annotations is available with InterMine and we were able to easily extend the parser to include the annotation method detail for each GO annotation.
The InterMine software also allows the flexibility of adding data types that are unique to SGD. The expression pattern of each gene is described in 352 microarray expression data sets provided at SGD with the SPELL software (ref. 6, http://spell.yeastgenome.org/). The addition of expression data required extending the data model to include expression-specific classes for genes like SpellDataSet, SpellDataSetCondition and SpellExpressionScore. The Gene class has been expanded to include the SpellExpressionScores class, which associates scores from many expression experiments with a gene. Every score corresponds to a SpellDataSetCondition class, which describes the experimental condition, and each condition comes from a SpellDataSet, which defines an experiment/study.
The addition of homology data from TreeFam (5) demonstrates YeastMine's flexibility at integrating data types that are not curated by SGD. Homology data from TreeFam provides homologs in eight species for each S. cerevisiae gene and integrating the TreeFam data into YeastMine required no customization.
Because all facets of the data (such as a gene, a GO term, an experimental condition, or a publication) are stored in YeastMine as entities known as Objects, these data become integrated when new data types share common Objects. GO annotations, homology and expression data all describe a gene (otherwise known as a gene Object in YeastMine), and so these data types are integrated with other data in YeastMine that have shared gene Objects during the weekly build. The build process allows these and any new data types to become incorporated with the majority of literature-curated data types including sequence and region information and phenotypes. This integrative data model allows researchers to query across multiple data types using YeastMine's powerful query interface.
Accessing data in YeastMine
Although SGD provides access to all its data types via multiple entry points (Table 1), it is difficult to query across multiple data types or easily search with a list of genes. Also, though it is possible to perform simple queries on some of the more complex data in SGD (e.g. GO annotations, expression levels) the richness of information inherent to these types of data beg for more sophisticated querying capabilities in order to examine all of the meaningful interpretations possible. Implementing these types of searches at SGD would require extensive modification of existing software or development of complicated custom query engine. However, within YeastMine these types of searches, such as retrieving GO annotations filtered for multiple criteria, or finding microarray data performed under a particular condition, become straightforward. YeastMine also differs from traditional web-based tools, which are limited to the options available on the interface, by offering a framework that gives the user the freedom to define their own parameters.
A YeastMine link is available at the top of all SGD pages and is available via the URL http://yeastmine.yeastgenome.org. The YeastMine homepage offers options to Search and Analyze data. The initial input can either be a query or a list of genes. Any type of data can be queried via Templates or the Query builder. Lists of data identifiers can be uploaded by the user, selected from a set of SGD-provided lists, or generated from the output of a query.
Templates
A Template is defined as a simple search interface for a predefined query. YeastMine provides a variety of templates that are grouped by data type. Each Template is shown with a short description of the search performed (http://yeastmine.yeastgenome.org/yeastmine/templates.do). Templates can be constrained to a default value or to a list of related data Objects. A majority of the templates are gene-centric, i.e. they allow for the retrieval of a particular data type for a gene Object or a list of gene Objects. The default gene Object in YeastMine includes all the feature types that are present in the gene association file (GAF) (Uncharacterized and Verified ORFs, pseudogenes, transposable element genes, RNAs and genes ‘Not in Systematic Sequence of S228C’). An example of a template search that retrieves a list of genes is the ‘Chromosome→Genes’ template. Using this template, the user selects the desired chromosome from a pull-down menu and the search retrieves all gene Objects from the chromosome of choice.
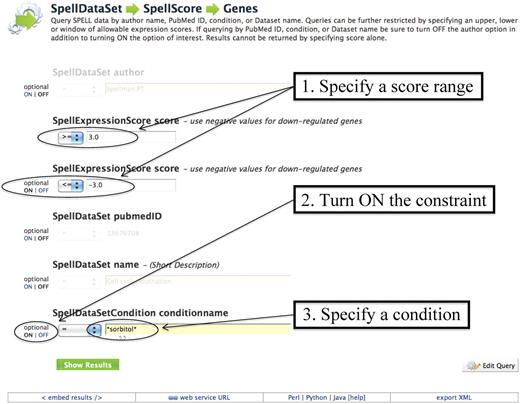
Another common template search will identify all genes whose expression is affected above a given cut-off score in response to osmotic stress. The ‘SpellDataSet→SpellScore→Genes’ template (Figure 1) provides an option to turn ON the ‘SpellDataSetCondition conditionname’ constraint. Entering the osmotic stress-inducing chemical ‘sorbitol’ into this field will search all expression data for those experiments performed under conditions that included this chemical. By selecting a score cut-off using the ‘SpellExpressionScore score’ option in the same template, the list of genes that is returned can be narrowed further. In addition to conditions and scores, it is possible to search for specific data sets, authors and publications using this template. Although one can view all of these data facets in the expression data tool SPELL (6), it is not possible to retrieve just this subset of genes through the current version of SPELL. By enabling the researcher to perform this expanded search through a predefined query, YeastMine is able to provide additional functionality for expression data analysis.
Example of a template search of expression data: screenshot of the ‘SpellDataSet→SpellScore→Genes’ template showing the SpellExpression Score constrained to be between ≥3 and ≤−3, and the SpellDatasetCondition name constrained to be ‘=*sorbitol*’. Switching ON the other parameters such as ‘SpellDataSet author’ or ‘SpellDataset pubmedID’ will allow constraint of those values. The ‘Show Results’ button runs the query. This template is prepopulated with certain constraints, but clicking on the ‘Edit Query’ button will bring up the Model browser, which offers more options for query constraints and output formats.
Lists
Another functionality that YeastMine adds to SGD is the ability to upload, query, retrieve, download and manipulate lists of different data types. Lists can be made for any Object entity as defined earlier in the ‘Creating YeastMine’ section of this paper, such as a list of genes or GO Term identifiers. They can be predefined by SGD, user-generated via uploading, or saved from the results of a query. The predefined lists include gene sets such as Verified ORFs, Uncharacterized ORFs, and tRNAs and are available from the ‘View’ submenu of the YeastMine Lists tab. Custom lists can be created through the ‘Upload’ submenu of the YeastMine Lists tab. Potential inputs for a custom list could be the result of a query at SGD or a list of genes identified in a genetic screen. Results from executed queries can be selected and added to a list via the ‘Create List’ option at the top of all search results.
Once a list is created, it can be used for additional queries or comparison with other lists. Lists can be used to restrict template queries to search for results relevant to that list. Templates where this option is available will have a ‘constrain in’ check box option that is followed by a pull-down menu populated by the SGD premade lists and any lists created by the user within their search session. Lists can be manipulated to perform functions such as joining lists, finding the intersection between lists, or subtracting lists to find features unique for some desired characteristics. In addition several widgets are available to analyze the lists further. The GO enrichment widget, for example, determines statistically significant enrichment of GO terms for a list of genes.
Query builder and Model Browser
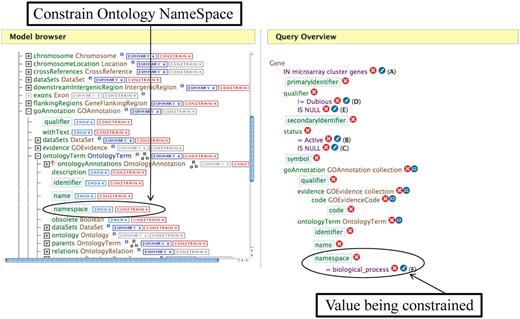
In addition to searching YeastMine with templates defined by SGD, it is possible to modify any existing query or template, or even to build one from scratch using the Query Builder function. In the Query Builder tab, the Model Browser displays the data present in YeastMine in an easy to navigate form and can be used to select and build a new or edit a predefined query. A new query can be built starting with any YeastMine data object such as Gene or GO annotation or Phenotype. The default Gene object in YeastMine mirrors the classifications of genes defined by SGD such as Verified ORFs, Uncharacterized ORFs, ‘Not in Systematic Sequence of S228C’. Similarly, the default Phenotype object mirrors all the attributes that are curated and displayed in the main SGD database. Users can modify any template using ‘Edit Query’ to customize data retrieval and display. A predefined query can also be edited using the Model Browser to include or exclude data or data attributes. Query Builder allows query customization by the ability to constrain on any Object, and choice of various data output options. This enables the user to build a custom query that suits their specific data search and retrieval needs. For example, if one has a list of genes that have correlated gene expression and would like to download the GO Biological Process annotations for those genes, it is fairly straightforward to modify an existing template to get these data. After saving the genes from a microarray cluster as a list using the List feature, one can go to the ‘Gene⋄GO terms’ template, restrict the query to use the saved list and then by editing the query using the Model Browser, add constraints to the Ontology Name Space to retrieve just the Biological Process annotations (Figure 2).
An example of editing a template using the Query Builder. The Model Browser (on the left) displays the attributes for the GOAnnotation object in the Gene→GO Terms template. Clicking on the ‘CONSTRAIN→’ button next to the namespace box allows one to constrain on the ontology namespace. The Query Overview (on the right) shows the ontology namespace being restricted to the value ‘Biological Process’.
Example of a complex query
The query described in detail in Table 2 illustrates the versatility of YeastMine to create a complex query to retrieve a list of GO protein complexes that have at least one member affected by osmotic stress. To identify these complexes, we first query the expression data to retrieve genes that are differentially expressed in response to osmotic stress. Next, we retrieve genes that have a mutant phenotype under osmotic stress conditions and finally we identify which of these genes have GO annotations indicating that they are constituents in macromolecular complexes. This example shows how to crossquery and combine the three different data types of gene expression, phenotype and GO annotation. In addition it highlights several features of YeastMine; combining lists, manipulating predefined templates and most importantly the ability to work with different data types using a single tool.
Step-by-step description of an intricate query using YeastMine to retrieve a list of protein complexes where one or more of the constituent members shows a response to osmotic stress
| Aim . | Template search . | Query builder edits . | List operations . |
|---|---|---|---|
| Step 1: retrieve genes differentially expressed in response to sorbitol (also shown in Figure 2) | SpellDataSet → SpellScore → Genes
| None | Save genes from results report as ‘List 1’ |
| Step 2: retrieve genes sensitive to osmotic stress when mutated | Phenotype → Genes
| None | Save genes from results report as ‘List 2’ |
| Step 3: retrieve genes sensitive to sorbitol when mutated | Phenotype → Genes | Query Overview
| Add genes from results report to ‘List 2’ |
Model Browser
| |||
| Step 4: make a list of all genes with a response to osmotic stress | None | None | Union ‘List 1’ and ‘List 2’ and Save as ‘List 3’ |
| Step 5: retrieve genes annotated with GO to a complex | GO Term name [and children of this term] → All genes in organism
| None | Save genes from results report as ‘List 4’ |
| Step 6: make a list of genes that respond to osmotic stress that are also in a complex | None | None | Intersect ‘List 3’ and ‘List 4’ and Save as ‘List 5’ |
| Step 7: retrieve complexes where at least one member protein responds to osmotic stress | Gene → GO term
| Model Browser
| Save GO child terms from results report as ‘End List’ |
| Aim . | Template search . | Query builder edits . | List operations . |
|---|---|---|---|
| Step 1: retrieve genes differentially expressed in response to sorbitol (also shown in Figure 2) | SpellDataSet → SpellScore → Genes
| None | Save genes from results report as ‘List 1’ |
| Step 2: retrieve genes sensitive to osmotic stress when mutated | Phenotype → Genes
| None | Save genes from results report as ‘List 2’ |
| Step 3: retrieve genes sensitive to sorbitol when mutated | Phenotype → Genes | Query Overview
| Add genes from results report to ‘List 2’ |
Model Browser
| |||
| Step 4: make a list of all genes with a response to osmotic stress | None | None | Union ‘List 1’ and ‘List 2’ and Save as ‘List 3’ |
| Step 5: retrieve genes annotated with GO to a complex | GO Term name [and children of this term] → All genes in organism
| None | Save genes from results report as ‘List 4’ |
| Step 6: make a list of genes that respond to osmotic stress that are also in a complex | None | None | Intersect ‘List 3’ and ‘List 4’ and Save as ‘List 5’ |
| Step 7: retrieve complexes where at least one member protein responds to osmotic stress | Gene → GO term
| Model Browser
| Save GO child terms from results report as ‘End List’ |
Retrieval of genes that have altered expression under osmotic stress, (Step 1, List 1); have a mutant phenotype under osmotic stress conditions (Steps 2–3, List 2). Lists created by Steps 1–3 are unified in Step 4 to obtain List 3. A list of genes (List 4) mapping up to the cellular component GO term ‘macromolecular complex’ is retrieved in Step 5. Intersecting List 3 and List 4 in Step 6 results in List 5, genes that both have a response to osmotic stress and are members of a complex. Finally, in Step 7, limiting our search to genes within List 5, we retrieve a list of GO complex terms that have at least one member of the complex experimentally shown to be involved in osmotic stress, the End List. The results of the End List using YeastMine version 2011-10-09 and using ‘3’ as an expression score cut-off in Step 1 can be found in Supplementary Table S1.
Step-by-step description of an intricate query using YeastMine to retrieve a list of protein complexes where one or more of the constituent members shows a response to osmotic stress
| Aim . | Template search . | Query builder edits . | List operations . |
|---|---|---|---|
| Step 1: retrieve genes differentially expressed in response to sorbitol (also shown in Figure 2) | SpellDataSet → SpellScore → Genes
| None | Save genes from results report as ‘List 1’ |
| Step 2: retrieve genes sensitive to osmotic stress when mutated | Phenotype → Genes
| None | Save genes from results report as ‘List 2’ |
| Step 3: retrieve genes sensitive to sorbitol when mutated | Phenotype → Genes | Query Overview
| Add genes from results report to ‘List 2’ |
Model Browser
| |||
| Step 4: make a list of all genes with a response to osmotic stress | None | None | Union ‘List 1’ and ‘List 2’ and Save as ‘List 3’ |
| Step 5: retrieve genes annotated with GO to a complex | GO Term name [and children of this term] → All genes in organism
| None | Save genes from results report as ‘List 4’ |
| Step 6: make a list of genes that respond to osmotic stress that are also in a complex | None | None | Intersect ‘List 3’ and ‘List 4’ and Save as ‘List 5’ |
| Step 7: retrieve complexes where at least one member protein responds to osmotic stress | Gene → GO term
| Model Browser
| Save GO child terms from results report as ‘End List’ |
| Aim . | Template search . | Query builder edits . | List operations . |
|---|---|---|---|
| Step 1: retrieve genes differentially expressed in response to sorbitol (also shown in Figure 2) | SpellDataSet → SpellScore → Genes
| None | Save genes from results report as ‘List 1’ |
| Step 2: retrieve genes sensitive to osmotic stress when mutated | Phenotype → Genes
| None | Save genes from results report as ‘List 2’ |
| Step 3: retrieve genes sensitive to sorbitol when mutated | Phenotype → Genes | Query Overview
| Add genes from results report to ‘List 2’ |
Model Browser
| |||
| Step 4: make a list of all genes with a response to osmotic stress | None | None | Union ‘List 1’ and ‘List 2’ and Save as ‘List 3’ |
| Step 5: retrieve genes annotated with GO to a complex | GO Term name [and children of this term] → All genes in organism
| None | Save genes from results report as ‘List 4’ |
| Step 6: make a list of genes that respond to osmotic stress that are also in a complex | None | None | Intersect ‘List 3’ and ‘List 4’ and Save as ‘List 5’ |
| Step 7: retrieve complexes where at least one member protein responds to osmotic stress | Gene → GO term
| Model Browser
| Save GO child terms from results report as ‘End List’ |
Retrieval of genes that have altered expression under osmotic stress, (Step 1, List 1); have a mutant phenotype under osmotic stress conditions (Steps 2–3, List 2). Lists created by Steps 1–3 are unified in Step 4 to obtain List 3. A list of genes (List 4) mapping up to the cellular component GO term ‘macromolecular complex’ is retrieved in Step 5. Intersecting List 3 and List 4 in Step 6 results in List 5, genes that both have a response to osmotic stress and are members of a complex. Finally, in Step 7, limiting our search to genes within List 5, we retrieve a list of GO complex terms that have at least one member of the complex experimentally shown to be involved in osmotic stress, the End List. The results of the End List using YeastMine version 2011-10-09 and using ‘3’ as an expression score cut-off in Step 1 can be found in Supplementary Table S1.
Results page/output formats
The ability to manage and download the results of a query in a convenient format is as important as being able to perform the query itself. The data columns in all of the result reports are customizable, enabling the user to choose exactly what type of information is in the output of a search. This feature is available both from the record results page and through the Query Builder. It is also possible to export all results either as a list for further querying within YeastMine, as a table to the Galaxy tool (7), or as a file to your desktop. YeastMine supports data download in multiple formats (tab delimited, comma separated, excel) and GFF3 format for sequence related data. We are working toward including other standard formats such as gene association file (GAF) format for GO annotations, and PSI MITAB and OSPREY for interaction data in future releases.
Additional features
In addition to enhancing SGD's data search and retrieval capabilities, additional features are available that facilitate the usability and individual customization of YeastMine. These include personalized search options, video tutorials, Web Services and interoperability between SGD and other Model organism databases (MODs).
MyMine
All queries and lists can be saved for use in future YeastMine sessions by creating a personalized ‘MyMine’ account. MyMine creates a private workspace for the user to create and save queries, templates and lists.
Tutorials and help pages
SGD has created a popular set of video tutorials to demonstrate the various features of YeastMine, to help users navigate the interface and to provide step-by-step directions through various search scenarios. These tutorials are linked from the YeastMine home and are directly available via the URL http://yeastgenome.org/video_tutorials.shtml#YMT. Detailed protocols outlining various use cases of YeastMine have been previously published (8). For questions not covered in the tutorials or protocols, emails to the SGD helpdesk (sgd-helpdesk at lists.stanford.edu) will result in a prompt response.
Web services
InterMine provides a RESTFUL API and WebServices thus allowing programmatic access to the mine. This allows bioinformatics and programming communities to use YeastMine as a data-mining tool. The Web Services feature has been utilized in the creation of a YeastGenome iPhone/iPad app that will be available in early 2012.
MOD interoperability
Another major benefit gained from utilizing the InterMine environment is the ability to interface with other MODs on a common platform. One aim of the InterMine project is not only to provide expanded querying capabilities within an individual MOD, but also to facilitate cross-organism querying between MODs. By integrating curated homology data from TreeFam, and in the future Ensembl Compara, InterMine provides a mechanism to easily navigate and query data from multiple organisms via their homologs. Several MODs and research projects have implemented the InterMine data warehouse system including: RGD (RatMine, http://ratmine.mcw.edu), ZFIN and modENCODE (http://InterMine.modencode.org/). Recently, work has begun on InterMine implementations at MGI and WormBase. See http://mods.InterMine.org/wiki/Interoperability for more details.
Future plans and conclusions
YeastMine enables querying across multiple data sources, navigating between data types and managing data all in one tool. YeastMine is being constantly improved and expanded to include more data types and include more functionality such as graphical data visualization, including a histogram that summarizes the expression levels of a gene in multiple data sets and graphs that summarize protein–protein and genetic interactions. Another future area of development focuses on integrating genomic sequence data describing regulatory regions and transcription in order to enable coordinate-based querying across multiple genomic features. New templates and data types are added to YeastMine on an ongoing basis and we welcome any questions regarding YeastMine or requests for new features at the SGD helpdesk. SGD is committed to being the primary source of S. cerevisiae data and will continue to provide its flagship tools and resources as well as striving to develop new services that meet the ever-evolving needs of our community.
Funding
National Human Genome Research Institute: Saccharomyces Genome Database project (grant number P41 HG001315); YeastMine development (grant number R01 HG004834, PI: to G.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health. Funding for open access charge: National Human Genome Research Institute (NHGRI) (P41 HG001315).
Conflict of interest. None declared.
Acknowledgements
We wish to acknowledge Richard Smith at University of Cambridge, UK for valuable assistance; members of the growing collaboration of MODs implementing mines; and the Saccharomyces Genome Database project staff that has provided testing, development of tutorial videos and thoughtful discussions to expand our use of YeastMine.