-
PDF
- Split View
-
Views
-
Cite
Cite
Kimberly Van Auken, Petra Fey, Tanya Z. Berardini, Robert Dodson, Laurel Cooper, Donghui Li, Juancarlos Chan, Yuling Li, Siddhartha Basu, Hans-Michael Muller, Rex Chisholm, Eva Huala, Paul W. Sternberg, the WormBase Consortium, Text mining in the biocuration workflow: applications for literature curation at WormBase, dictyBase and TAIR, Database, Volume 2012, 2012, bas040, https://doi.org/10.1093/database/bas040
Close - Share Icon Share
Abstract
WormBase, dictyBase and The Arabidopsis Information Resource (TAIR) are model organism databases containing information about Caenorhabditis elegans and other nematodes, the social amoeba Dictyostelium discoideum and related Dictyostelids and the flowering plant Arabidopsis thaliana, respectively. Each database curates multiple data types from the primary research literature. In this article, we describe the curation workflow at WormBase, with particular emphasis on our use of text-mining tools (BioCreative 2012, Workshop Track II). We then describe the application of a specific component of that workflow, Textpresso for Cellular Component Curation (CCC), to Gene Ontology (GO) curation at dictyBase and TAIR (BioCreative 2012, Workshop Track III). We find that, with organism-specific modifications, Textpresso can be used by dictyBase and TAIR to annotate gene productions to GO's Cellular Component (CC) ontology.
Introduction
Biocuration is the collection and organization of biological data into machine-readable forms that can be stored in databases and presented to scientists, largely through the World Wide Web. The past 15–20 years have seen a tremendous increase in the number of organism-specific or data type–specific databases available to the scientific community (1). Such databases typically rely on manual curation of the primary scientific literature for their content.
Although manual curation is thorough and captures many critical experimental details, it is slow and efforts to improve the efficiency of manual curation are needed (2). Progress towards improving the rate of manual curation requires an understanding of curation workflow so that useful tools can be implemented at key steps in the curation pipeline (3). Although the specific requirements of manual curation may vary somewhat between groups, some general principles do apply. Most groups need to (i) find potentially relevant documents for curation, (ii) determine what data types or experiments are contained within those documents (a process varyingly known as triage, flagging or first-pass curation), (iii) identify the specific entities to which biological knowledge will be assigned (entity recognition) and (iv) extract experimental information from the full text and convert it into a machine-readable form for database entry.
WormBase (http://www.wormbase.org) is a model organism database that curates data about Caenorhabditis elegans and other nematodes (4). Although WormBase is largely a gene-centric database, it warehouses, in addition to genomic sequence and gene function curation, information about additional aspects of nematode biology, including anatomy, reagents, researchers and publications. Currently, WormBase releases a new version of the database six times a year.
In the past, nearly all WormBase literature curation was performed as a fully manual process. Over time, however, we have incorporated a number of text-mining approaches into our curation pipeline, allowing us to transition to a more automated workflow. Specifically, use of support vector machines (SVMs) (5), entity recognition scripts and the Textpresso information retrieval system (6) has all contributed to our increasingly automated approach.
In this article, we describe the WormBase curation workflow as presented at the Critical Assessment of Information Extraction Systems in Biology (BioCreative) 2012 Workshop Track II on Workflow (http://www.biocreative.org). In addition, we describe the application of one aspect of the WormBase curation pipeline, using the Textpresso information retrieval system for Gene Ontology (GO) Cellular Component Curation (CCC) (6,7), to the curation of subcellular localization data for two additional organisms, the social amoeba Dictyostelium discoideum, the biology of which is captured in dictyBase (8) (http://dictybase.org), and the flowering plant Arabidopsis thaliana, data for which is curated in The Arabidopsis Information Resource (TAIR) (9) (http://arabidopsis.org). This latter work was presented as part of the BioCreative 2012 Workshop Track III on Interactive Text Mining. Evaluation of Textpresso search results for Dictyostelium and Arabidopsis indicates that Textpresso can be used by dictyBase and TAIR to annotate gene products to GO's CC ontology.
Track II: WormBase workflow
General overview
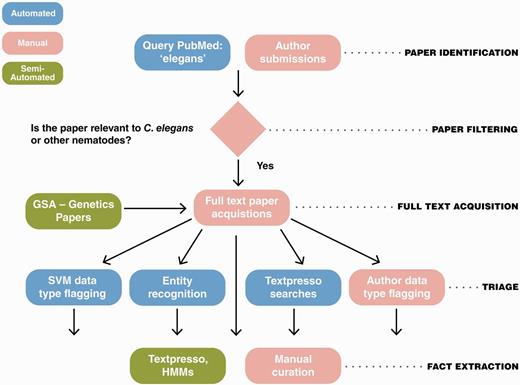
The WormBase literature curation workflow is given in Figure 1. Briefly, articles relevant to WormBase curation are typically identified through automated PubMed searches and then processed through automated and manual triage methods. Text mining and manual curation are subsequently used for fact extraction. Each of these steps is described in more detail below.
The WormBase literature curation workflow. WormBase literature curation incorporates automated (blue), semi-automated (green) and manual (pink) steps. Potentially curatable papers are initially brought into WormBase primarily via PubMed searches with additional contributions from authors and a collaboration between WormBase and the GSA for Genetics and G3: Genes, Genomes, Genetics papers. Following full-text acquisition, a triage step is used to determine what data types are present in papers. The triage step is largely automated but also includes author contributions. Once papers have been flagged for data types, curators responsbile for curation of that data type use manual and semi-automated methods to extract the information and convert it into machine-readable format.
Paper identification and filtering: PubMed queries
Papers curated for WormBase typically enter the curation pipeline through an automated, daily PubMed search using the keyword ‘elegans’. This search, performed by a Perl script, identifies newly submitted papers that contain the keyword ‘elegans’ in the title or abstract, as well as papers submitted at an earlier date that did not contain the keyword in the title or abstracts but have been indexed using the MeSH term Caenorhabditis elegans.
Bibliographic information from papers identified by the PubMed search is presented to a curator for manual approval in a Web-based form. The form lists the PubMed identifier, the authors, abstract, journal and PubMed publication type. In most cases, papers can be approved or rejected based on the content of the abstract, but in some cases, curators need to access the full text of the paper before making a final decision. Accepted papers are also given a designation of ‘primary’ or ‘not primary’ to indicate whether the paper is likely to contain primary experimental data. The primary/not primary designation can help curators prioritize papers for curation. Currently, WormBase identifies ∼1200 papers per year for curation.
Paper identification and filtering: author submission
WormBase also receives papers directly from authors. Most often these papers are from journals not currently indexed by PubMed. For author-submitted publications, a curator assesses the relevance of the paper to WormBase before accepting the paper. Bibliographic information for author-submitted papers is manually entered into the curation database.
Paper full-text acquisition
WormBase strives to download the full text of every paper approved by us for inclusion in the curation database. The full text of papers, including supplementary material, is downloaded manually from journal websites and provides the input for all subsequent text mining and curation.
Paper identification and full-text acquisition: WormBase collaboration with the Genetics Society of America
In collaboration with the Genetics Society of America (GSA) and Dartmouth Journal Services, WormBase receives pre-publication access to C. elegans articles accepted by the journals Genetics and G3: Genes, Genomes, Genetics. The full text of these articles, and their corresponding Digital Object Identifiers, are initially submitted to WormBase from the GSA through a Web service with full bibliographic information subsequently retrieved through the PubMed pipeline. Papers published in Genetics and G3: Genes, Genomes, Genetics articles are curated, in part, through a text markup pipeline that adds hyperlinks to entities within the paper to WormBase Web pages, a process that can also serve as a curation flagging mechanism (10). For example, as part of this pipeline, authors can indicate whether their paper describes new entities, such as genes, variations and strains, for which WormBase Web pages do not yet exist.
Triage overview
Triage is a key step in the biocuration pipeline and, broadly speaking, involves identification of specific data types for curation. Entity recognition is equally important and involves identification of specific objects to which biological knowledge, such as functional annotation or sequence, may be associated. Before 2009, WormBase literature triage was a fully manual process. Since that time, however, we have transitioned to a semiautomated triage process that involves use of a number of different approaches described below.
Triage: data type flagging using support vector machines
SVMs, a type of supervised learning method, are currently used at WormBase to ‘flag’ papers for 10 different data types: expression patterns, antibodies, genetic interactions, physical interactions, RNAi phenotypes, variation phenotypes, overexpression phenotypes, gene regulation, gene structure (model) corrections and variation sequence changes. For each data type, SVM models were trained using known positive and negative documents, that is previously curated or flagged papers. Selection of data types for SVM analysis was based on both availability of suitable training sets and curation priorities. The training sets typically included several hundred to over a thousand positive documents (depending on the frequency of the data type in the C. elegans literature) and a few thousand negative documents.
Currently, SVM analyses using a nine-component comprehensive scheme (i.e. nine different SVM models) are performed biweekly on the full text of newly acquired papers. SVM results are presented to curators on a Web page that lists predicted positive and negative papers. Positive papers are further classified as being of high, medium or low confidence, an empirical confidence measure based on the number of SVMs in the comprehensive scheme that yield a positive prediction (i.e. 7–9 models = high, 4–6 models = medium and 1–3 models = low). Details on the methods used for training and testing the SVMs currently in use have been published elsewhere (5). Ongoing maintenance involves monitoring performance by curator feedback on false-positive and -negative papers and periodic retraining of the algorithm with updated training sets.
Triage: entity recognition and data type flagging using Textpresso
The Textpresso information retrieval system (http://textpresso.org) is also used for entity recognition and data type flagging. Textpresso is an information retrieval and extraction system that uses keyword and/or category searches on the full text of papers to identify sentences within documents that match search criteria (6). Categories are ‘bags of words’ that encompass terms of a common semantic concept. Searches can be restricted to specific paper sections, maximizing the likelihood that identified sentences are relevant to experimental results reported in the paper.
WormBase curators employ both manual and automated Textpresso searches on the full text of papers to identify documents containing entities or data types of interest. Recognition of biological entities in full text, such as variations, transgenes and molecules, is an essential aspect of WormBase curation. In cases where the entity in question conforms to standard C. elegans nomenclature, such as variations, and transgenes, pattern matching using regular expressions and Perl scripts is employed for entity identification. In other cases, for example molecules, lists of entities mined from databases such as ChEBI (11) are used.
Curators may also use Textpresso category searches for data type flagging. If necessary, curators design new, curation task-specific Textpresso categories to perform these searches. For example, one recently developed Textpresso-based pipeline flags papers that mention the C. elegans homolog of a human disease gene or a C. elegans model of a human disease. These searches employ Textpresso categories of C. elegans gene names, human disease terms and keywords such as ‘ortholog’, ‘homolog’, ‘model’ and ‘similar’. The Textpresso human disease category incorporates terms from three different sources: (i) the Neuroscience Information Framework (12), (ii) the Disease Ontology (13) and (iii) the Online Mendelian Inheritance in Man (14), with iterative modifications made to optimize search results for the C. elegans literature.
Triage: author flagging
Although nearly all data types have been included in either an SVM or a Textpresso-based triage pipeline, WormBase also employs an author flagging pipeline, similar to that used at FlyBase (15), to encourage authors to manually flag data types present in their recently published papers. Corresponding authors are contacted through email shortly after their publication is incorporated into the WormBase curation database. The email message contains a link to a form where authors can flag the data types in their paper and, optionally, provide experimental details. Over the past 2 years, the response rate for this pipeline has been 40%, with ∼75% of respondents supplying additional details beyond simply flagging yes or no. Within that 75%, we find that two-thirds of the respondents supply data beyond simply listing the genes studied in the paper, with additional information on mutant phenotypes, gene site of action (e.g. tissue or cell type) and genetic interactions added most frequently. The accuracy (i.e. precision or measure of false-positive rate) of author flagging varies with data type. Data types such as RNAi experiments, expression patterns and chemicals are flagged with >97% precision, whereas data types such as variation and overexpression phenotypes are flagged with 90 and 87% precision, respectively, and anatomy function and gene product interactions are flagged with 71 and 62% precision, respectively.
Fact extraction overview
Once papers have been flagged for different data types, curators are tasked with extracting the data in a manner that conforms to the data models of the main WormBase curation database, acedb (http://www.acedb.org/). Curators extract experimental details via either fully manual curation or semiautomated methods that incorporate the results of Perl scripts, Textpresso category searches or Hidden Markov Models (HMMs).
Broadly speaking, WormBase curation consists of creating and characterizing entities (e.g. creating a database object for a newly described variation and its associated sequence change) or linking two or more entities in a biological relationship, often using controlled vocabularies (e.g. gene A regulates gene B with respect to process P in cell type C). Free-text descriptions are also used, for example, to create concise gene function descriptions or capture experimental details.
Fact extraction: manual fact extraction
In most cases, WormBase literature curators manually enter data into a curation database using Web-based forms. One form, the Ontology Annotator (OA), is used to curate 14 different data types. The OA is based on the Phenote (http://phenote.org/) curation tool developed by the Berkeley Bioinformatics Open-Source Projects (http://berkeleybop.org/) for the purposes of annotating biological phenotypes using ontologies. The key features of the OA, in addition to easily annotating using ontologies, include autocompletion of searches, drop-down menus for short lists, the ability to view information about other curated objects and error checking. Information entered through the OA is stored in a PostgreSQL database maintained in a Linux operating system environment. Prior to each build of the WormBase database, data are exported from the PostgreSQL database in file formats conforming to the acedb database, which underlies much of the WormBase website.
Fact extraction: semiautomated fact extraction using Textpresso category searches and HMMs
In addition to fully manual fact extraction, WormBase curators also use the results of Textpresso category searches for fact extraction in several semiautomated curation pipelines. In some cases, the Textpresso searches are performed on a subset of SVM-positive papers to help prioritize high-confidence papers for curation. Textpresso-derived sentences describing, for example, genetic interactions and subcellular localization (GO CCC) are presented in the context of curation tools for curators to review and use for fact extraction. For GO CCC, the curation entity and, where possible, suggested annotations based on previous curation are pre-populated on the form (7). Pre-populating data fields on the curation form saves curator time and leverages previous curation for new annotation, a feature that can also serve as an internal annotation consistency check.
Currently, HMMs are also used at WormBase to curate enzymatic and transporter activities to GO’s molecular function (MF) ontology. SVMs and sophisticated keyword and category searches have some difficulties recognizing entities and facts on a sentence or paragraph level because they take limited account of the context in which an entity is used and do not take advantage of the sequential nature of sentences. However, HMMs are well suited to alleviate these drawbacks.
We applied an HMM method to GO MF curation by using 200 sentences describing enzymatic and transporter activities to train the HMM. The procedure involved selection of a list of feature words, processing of individual sentences using these features and then training and testing the resulting feature sequences using HMMs. The feature words were selected according to their frequency in the training sentences compared with a background frequency computed from the entire C. elegans corpus. The relative threshold for including overrepresented words as features was determined by testing the resulting HMM on a test set and optimizing the F-score. We then scanned the entire C. elegans corpus for sentences describing enzymatic and transporter activities to make new annotations. For this curation task, curators use a Web form that lists sentences from the full text of papers ranked according to the probability that they describe the result of an enzymatic or transporter assay.
Although for the purposes of describing the pipeline, triage, entity recognition and fact extraction are represented as three separate aspects of our curation workflow, in practice, there is some overlap between these pipelines. For example, an entity recognition script that identifies a newly described variation may simultaneously flag the paper in which the variation is reported for variation phenotype curation. Likewise, Textpresso category searches that identify sentences describing subcellular localization simultaneously flag the paper for expression pattern information. A table describing the different data types curated at WormBase and the various methods used for their curation is available at http://wiki.wormbase.org/index.php/WormBase_Literature_Curation_Workflow.
Encoding methods: entities, relationships and their representation in the database
Data in WormBase are represented as distinct classes that store information about specific instances of that class. Examples of classes include genes, molecules, RNAi experiments and interactions. Each class has a corresponding acedb data model that organizes the information needed to accurately represent that class in the database. Information captured in the data model may be relatively simple, such as an external database identifier, or it may be more complex, such as the details of an RNAi experiment. Evidence (e.g. a publication) for the information contained within a data model may be associated directly with the information in the data model or may instead be found in another database object to which the original model refers.
Encoding methods: use of standardized and controlled vocabularies
WormBase curators use a number of ontologies, including the GO (16), Sequence Ontology (17), Worm Phenotype Ontology (18), Cell and Anatomy Ontology (19) and Life Stage Ontology (19), for data type curation. Internal controlled vocabularies are also used to capture, for example, details about antibody production such as the organism in which an antibody was produced and whether the antibody is tissue specific.
Information access: curation difficulties and their resolution
Curation difficulties arise in two general ways. First, information presented in a publication may not be sufficient to link the data unambiguously to a WormBase database object. For example, curation of RNAi experiments requires mapping the sequence used as a reagent to the genomic sequence of the target. If the sequence of the reagent is not reported in a paper, curators may need to contact authors to request the necessary information. If no further information is available, the curated data may be assigned to the most general entity possible, in this case the entire gene, or alternatively may not be captured in WormBase.
Second, information presented in a paper may not be sufficient for curators to confidently assign annotations without additional background knowledge. In these cases, curators may need to consult previous publications, WormBook (20) or additional online resources such as the GO website or Wikipedia (http://www.wikipedia.org) to assign the correct annotation.
Track III: Textpresso for GO cellular component curation at dictyBase and TAIR
Overview
dictyBase and TAIR are model organism databases for the social amoeba D. discoideum and related species and the flowering plant A. thaliana, respectively. Like WormBase, dictyBase and TAIR are members of the GO Consortium and annotate gene products to the three GO ontologies: Biological Process: biological process (BP), MF and cellular component (CC).
Recently, we began collaborating with dictyBase and TAIR to implement a version of the Textpresso information retrieval and extraction system for their literature corpora. As part of this collaboration, we wished to evaluate how the Textpresso for CCC pipeline implemented at WormBase (7) could be used to aid Dictyostelium and Arabdisopsis GO curation. Evaluation of CCC for dictyBase was presented as part of the BioCreative Track III session on Interactive Text Mining, with similar evaluation for Arabidopsis performed subsequent to the workshop. The details of the implementation and evaluation results for each group are presented below.
Implementation of Textpresso for dictyBase and TAIR
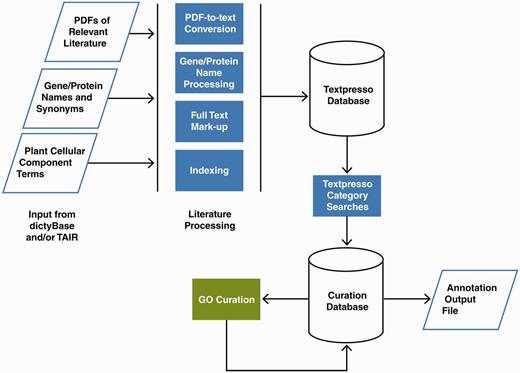
The Textpresso information retrieval and extraction system searches the full text of articles using keywords and/or categories containing semantically related terms. Key steps in implementing Textpresso for dictyBase and TAIR, illustrated in Figure 2, were (i) establishing a pipeline for paper acquisition, (ii) creating organism-specific categories for gene and protein names and (iii) fine tuning existing Textpresso categories to optimize search results for a more diverse group of organisms.
Pipeline for Textpresso for Cellular Component Curation for dictyBase and TAIR. PDFs of publications included in the dictyBase and TAIR curation corpora and files of Dictyostelium and Arabidopsis gene and protein names and synonyms are uploaded to a Textpresso server at Caltech. PDFs are converted to text; gene and protein names and synonyms are processed to include variants (e.g. upper- and lower-case versions), and organism-specific terms are added to the Textpresso cellular component ontology. Full text is then marked up using the new categories. Four- and five-category searches are performed on the full text and results formatted and stored for use in the curation database. Using a Web-based curation form, curators make annotations that are subsequently stored in the curation database and available for export as annotation files.
Paper acquisition for dictyBase and TAIR
To regularly acquire the full text of articles for dictyBase and TAIR, we established the following protocol. dictyBase and TAIR curators specify the publications that are relevant to their database curation pipeline, and then Textpresso processes the associated PDFs for full-text markup and indexing. For TAIR, there are ∼4000 new papers per year (∼2000 of which contain information for TAIR curation); for dictyBase, serving a smaller community, there are ∼200 publications per year.
Updating Textpresso categories for cellular component curation for dictyBase and TAIR
Textpresso for CCC was initially designed to retrieve sentences describing subcellular localization of C. elegans gene products from the full text of papers. To identify these sentences, papers are searched using four Textpresso categories: (i) assay terms (e.g. GFP, reporter, expression), (ii) verbs (e.g. expressing, detected, localizes), (iii) cellular component terms (e.g. nucleus, cytoplasmic, plasma membrane) and (iv) gene product names (e.g. DAF-16, COSA-1, LIN-17). Matching sentences must contain at least one term from each of these categories. For new implementations, we thus needed to make sure that each of the four categories was appropriate for the respective bodies of literature.
From curatorial experience, we reasoned that two of the categories, assay terms and verbs, were unlikely to differ significantly among the Dictyostelium, Arabidopsis and C. elegans literature, so initially we did not make any changes to these categories. However, the initial Textpresso cellular component category was derived solely from the C. elegans literature and thus lacked plant-specific terms such as chloroplast or thylakoid and included some terms that were not relevant to the Arabidopsis literature. To address any deficiencies, we revised the initial Textpresso cellular component category to include plant-specific terms and their plural forms and also included all macromolecular complexes as represented in the GO CC ontology. To address potentially irrelevant terms, we removed component terms such as process or processes, which are used to describe neuronal projections in C. elegans but do not correspond to a cellular component in plants. This revised Textpresso cellular component category was used for the dictyBase and TAIR evaluations.
Finally, the Textpresso for CCC searches require that a sentence describing subcellular localization contain the name of the gene product for which the experimental result is reported. The initial lists of genes, gene products, their synonyms and database identifiers were supplied by dictyBase and TAIR. In collaboration with both groups, we reviewed the existing dictyBase and TAIR gene name categories to ensure that all known forms of a gene and its encoded products (e.g. case variants) were included in this category. Also, for Arabidopsis genes preceded by the abbreviation ‘At’, such as AtMC9, we included both the full name (AtMC9) and the name without the ‘At’ (i.e. MC9) in the searches. In addition, on review of the gene name category, we decided to exclude some terms, such as actin for Dictyostelium, or gene symbols, such as ‘ER’ for Arabidopsis, as these names do not afford sufficient specificity for the purposes of our searches.
Evaluation of Textpresso for cellular component curation at dictyBase and TAIR
To evaluate the performance of Textpresso for CCC at dictyBase and TAIR, we compared a gold-standard set of sentences and annotations with those derived from Textpresso searches. In each case, gold-standard sentences describing subcellular localization and any associated annotations were selected by an experienced GO curator from 15 previously uncurated papers. Papers were selected from recent (i.e. 2011 or 2012) publication years and, as we wished to evaluate the performance of Textpresso for CCC, on the basis of whether they contained subcellular localization data. For dictyBase, 12 of the 15 papers contained subcellular localization data; the remaining three papers were true negative papers included in the evaluation set to help assess search precision. For TAIR, 13 of the 15 papers contained subcellular localization data; the remaining two papers were true negatives. For the dictyBase evaluation, two different curators, one from dictyBase and the other from the Plant Ontology project (21), both relatively newer to GO curation, participated in the evaluation. For the TAIR evaluation, an experienced GO curator performed the evaluation.
Evaluation strategy: Textpresso-based curation platform
To compare Textpresso searches to that of fully manual curation, the results of two different Textpresso searches were evaluated. The first search used the standard four categories, whereas the second search included a fifth category called Tables and Figures. The Tables and Figures category contains terms such as Figure, Fig and Table, and thus was used to identify only those statements that refer to a Figure or Table in the paper. We included the Tables and Figures category in the evaluation to determine whether restricting Textpresso searches to only those sentences that specifically reference a Figure or Table was sufficient for curation and what effect a more restrictive search would have on annotation metrics.
Each sentence returned by the Textpresso system receives a score based on the number of matching keyword or category terms. From previous experience, we find that true-positive sentences generally score between a range of 4 (i.e. one match to each of the four categories) and 20 and that sentences with a score >40 are nearly always false positives returned due to a high number of matches to terms from a list or table within the paper. Therefore, all Textpresso sentences were included in the evaluation with the exception of those sentences that received a Textpresso search score >40.
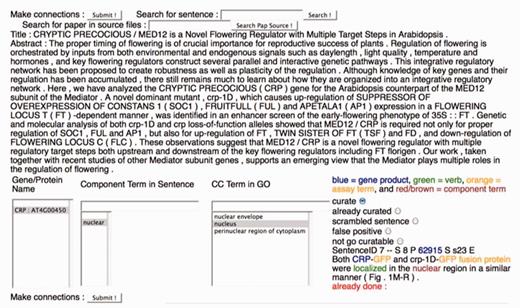
To perform the evaluation, curators used a Web-based annotation form (Figure 3). For the dictyBase evaluation, the form uniquely assigned annotations to each respective evaluator. Before performing the evaluation, curators received a brief tutorial on how to use the curation form. In the curation form, sentences identified through the Textpresso searches were displayed on the right side of the form, with terms from the four categories, Genes, Cellular Components, Assay Terms and Verbs, colour coded to indicate from which category the term match came. Curators used the columns on the left side of the form, ‘Gene/Protein Name’, ‘Component Term in Sentence’ and ‘CC Term in GO’ to make annotations. To speed up the curation process, the values for the ‘Gene/Protein Name’ and ‘Component Term in Sentence’ columns were pre-populated using the Textpresso output. Similarly, wherever possible, values for the CC Term in GO column were pre-populated based on previous GO curation (by WormBase and TAIR, see below).
The Textpresso for CCC Curation Form. A screenshot of the curation form used for the Textpresso for CCC evaluation is shown. Textpresso sentences are displayed on the bottom right corner of the form, with matches to each of the Textpresso categories highlighted and color coded. The title and abstract of the paper are shown at the top. On the bottom left side of the form are three curation boxes containing, from left to right, the identified gene product, the component term from the retrieved sentence and suggested GO terms based on the previous curation. To make a GO annotation, the curator makes a selection from each of the boxes, highlighted in gray, selects the curate radio button above the sentence and presses Submit to commit the annotation to the curation database. Additional radio buttons allow curators to further classify sentences, if needed. These additional actions were not part of the current BioCreative evaluation.
To make an annotation, the curator selected a gene or protein name (mapped to a database identifier for unambiguous annotation) in Column 1, the component term in the sentence in Column 2 and then a suggested GO term in Column 3. If an appropriate GO term was not suggested, then the curator entered a new GO term in Column 3. Suggested GO annotations arise from a relationship index, stored in a curation database, that links the component term selected in Column 2 to the GO term entered in Column 3. If a new GO term has been added to Column 3, a new relationship between the component term selected in Column 2 and the GO term entered in Column 3 is added to the index for future curation.
Evaluation metrics: sentences, annotations and curation efficiency
Textpresso search results were evaluated on three levels: (i) sentence retrieval, (ii) annotations made and (iii) estimated effects on curation efficiency. For analyzing sentence retrieval, we used metrics of precision and recall in which precision is defined as the percentage of sentences retrieved by Textpresso that were relevant (i.e. described subcellular localization) and recall is defined as the percentage of relevant sentences Textpresso retrieved from the test documents. For assessing annotation metrics, we adapted the standard definitions of recall and precision, defining precision as the percentage of annotations made from Textpresso sentences that exactly match the annotation in the gold-standard set and recall as the percentage of annotations made from Textpresso sentences that either exactly match or are a parent term of the gold-standard annotation. The recall metric reflects our assumption that biologically accurate annotations, even if they are of lesser granularity, are still potentially valuable for databases, especially in the context of semiautomated or fully automated curation pipelines. The F-score is also reported as an indication of the accuracy of the test. For analyzing Textpresso for CCC metrics, it is important to note that currently, the search criteria are designed to retrieve all sentences that describe subcellular localization of a gene product. Therefore, it is possible that the Textpresso searches will return sentences that, while describing subcellular localization, would not typically lead to a GO annotation. For example, a sentence that describes localization of a gene product in a non–wild-type background would be returned by Textpresso but would not be annotated for GO.
A summary of sentence retrieval metrics is presented in Table 1. For the dictyBase evaluation, the four-category Textpresso search identified sentences with a precision of 77.5% and recall of 37.9% (F-score: 50.9%). For the five-category dictyBase search, Textpresso identified sentences with a precision of 81.5% and a recall of 39.7% (F-score: 53.4%). For the TAIR evaluation, the four-category Textpresso search identified sentences with a precision of 57.5% and recall of 52.2% (F-score: 54.7%). For the five-category TAIR search, Textpresso identified sentences with a precision of 89.3% and a recall of 56.8% (F-score: 69.4%).
Results of Textpresso sentence retrieval for four- and five-category searches for Dictyostelium discoideum and Arabidopsis thaliana literature
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum | 0.379 | 0.775 | 0.509 | 0.397 | 0.815 | 0.534 |
| Arabidopsis thaliana | 0.522 | 0.575 | 0.547 | 0.568 | 0.893 | 0.694 |
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum | 0.379 | 0.775 | 0.509 | 0.397 | 0.815 | 0.534 |
| Arabidopsis thaliana | 0.522 | 0.575 | 0.547 | 0.568 | 0.893 | 0.694 |
Results of Textpresso sentence retrieval for four- and five-category searches for Dictyostelium discoideum and Arabidopsis thaliana literature
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum | 0.379 | 0.775 | 0.509 | 0.397 | 0.815 | 0.534 |
| Arabidopsis thaliana | 0.522 | 0.575 | 0.547 | 0.568 | 0.893 | 0.694 |
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum | 0.379 | 0.775 | 0.509 | 0.397 | 0.815 | 0.534 |
| Arabidopsis thaliana | 0.522 | 0.575 | 0.547 | 0.568 | 0.893 | 0.694 |
A summary of the annotation metrics is presented in Table 2. The two curators performing the dictyBase evaluation were able to make annotations from the four-category search with precision of 78.3 and 77.8% and recall of 37.1 and 14.5%, respectively (F-scores: 50.3 and 24.4%, respectively). For the five-category search, the curators were able to make annotations with precision of 75.0 and 71.4% and recall of 32.2 and 11.3% (F-scores: 45.0 and 19.5%, respectively). For the TAIR evaluation, the curator was able to make annotations from the four-category search with precision of 92.0% and recall of 46.0% (F-score: 61.3%) and annotations from the five-category search with precision of 91.3% and recall of 42.0% (F-score: 57.5%). These results reveal that, with respect to annotations, there was little difference between the four- and five-category Textpresso searches, with both annotation recall and annotation precision generally lower for the more restrictive five-category search.
Results of Textpresso-based cellular component annotation for four- and five-category searches for Dictyostelium discoideum and Arabidopsis thaliana literature
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum curator 1 | 0.371 | 0.783 | 0.503 | 0.322 | 0.750 | 0.450 |
| Dictyostelium discoideum curator 2 | 0.145 | 0.778 | 0.244 | 0.113 | 0.714 | 0.195 |
| Arabidopsis thaliana curator 1 | 0.460 | 0.920 | 0.613 | 0.420 | 0.913 | 0.575 |
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum curator 1 | 0.371 | 0.783 | 0.503 | 0.322 | 0.750 | 0.450 |
| Dictyostelium discoideum curator 2 | 0.145 | 0.778 | 0.244 | 0.113 | 0.714 | 0.195 |
| Arabidopsis thaliana curator 1 | 0.460 | 0.920 | 0.613 | 0.420 | 0.913 | 0.575 |
Results of Textpresso-based cellular component annotation for four- and five-category searches for Dictyostelium discoideum and Arabidopsis thaliana literature
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum curator 1 | 0.371 | 0.783 | 0.503 | 0.322 | 0.750 | 0.450 |
| Dictyostelium discoideum curator 2 | 0.145 | 0.778 | 0.244 | 0.113 | 0.714 | 0.195 |
| Arabidopsis thaliana curator 1 | 0.460 | 0.920 | 0.613 | 0.420 | 0.913 | 0.575 |
| . | Four-category search . | Five-category search . | ||||
|---|---|---|---|---|---|---|
| . | Recall . | Precision . | F-score . | Recall . | Precision . | F-score . |
| Dictyostelium discoideum curator 1 | 0.371 | 0.783 | 0.503 | 0.322 | 0.750 | 0.450 |
| Dictyostelium discoideum curator 2 | 0.145 | 0.778 | 0.244 | 0.113 | 0.714 | 0.195 |
| Arabidopsis thaliana curator 1 | 0.460 | 0.920 | 0.613 | 0.420 | 0.913 | 0.575 |
An additional aspect of the BioCreative Task III evaluation was an assessment of the amount of time it takes to perform the assigned curation task manually versus the amount of time it takes using the Textpresso system. For one dictyBase evaluator, the curator recorded that the Textpresso system resulted, overall, in an ∼2.5-fold increase in curation efficiency. For the second dictyBase evaluator, the curator noted no increase in efficiency with the first set of search results (five-category search), but after evaluating the second set of search results (four-category search), also recorded an ∼2.5-fold increase in curation efficiency, perhaps reflecting increasing familiarity with the organism and the curation system. For the TAIR evaluation, we instead compared the time spent manually curating the gold-standard set of annotations with the time spent annotating the same papers using Textpresso and found an ∼10-fold decrease in the amount of time spent in annotating when using Textpresso.
Analysis of false-negative and false-positive Textpresso results
As with the C. elegans searches (7), there are several different reasons for false-negative sentences in the dictyBase and TAIR results. In some cases, terms or variants of existing terms were missing from one of the Textpresso categories. For example, the current version of the Textpresso celluar component category contains the phrase ‘microtubule organizing center’, while the version of that phrase used in one of the evaluation papers was ‘microtubule-organizing center’. In another example, a paper repeatedly referred to protein localization to the EHM, an abbreviation for the extrahaustorial membrane, but that abbreviation is neither in the GO CC ontology nor the Textpresso cellular component category.
In other false-negative cases, the statement in a paper that described localization did so using only three of the four Textpresso categories, lacking, for example, a term from the assay category or using anaphora such as ‘it’ or ‘this protein’ to refer to a gene product described at an earlier point in the paper. Other reasons for false negatives include technical issues with correctly identifying gene names, such as those that contain Greek characters.
False-positive sentences returned by Textpresso also show common themes among the different implementations. As described previously for C. elegans (7), one source of false-positive results stems from run-on sentences that arise during the PDF-to-text conversion step that is part of the Textpresso software pipeline. In these cases, a subset of matching category terms is found across two or more sentences, neither of which fully expresses the information of interest. In other cases, false-positive sentences describe an aspect of a subcellular organelle in the organism studied but do not actually discuss localization of a gene product to that organelle.
Discussion
Biocuration has become an essential aspect of biological knowledge dissemination. Most biocuration is still performed manually by highly trained curators tasked with converting published reports into machine-readable data that can then be displayed, queried and mined via publically available websites. Complete, or even near-complete, curation of the existing biomedical literature will require tremendous manual effort and is unlikely to be finished in the foreseeable future. As effective use of biological knowledge requires information of both depth and breadth, there is a tremendous need to advance the utility of text-mining and natural language processing tools to improve the efficiency with which biocuration is performed.
WormBase curation workflow
Since its inception, the WormBase curation workflow has progressed from a nearly completely manual pipeline to one that increasingly incorporates automated and semiautomated procedures. For example, data type flagging, once a fully manual step, is now automated for 10 different data types using SVMs. In addition, some fact extraction pipelines, such as those used for curating genetic and physical interactions as well as GO CC and MF annotations, rely on semiautomated approaches that use Textpresso category searches or HMMs. Periodic assessment of metrics for each approach will allow WormBase curators to fine-tune these methods as we anticipate that both the content of the published literature and curation priorities may change.
In addition, WormBase collaborations with professional societies and the journals they publish, such as the GSA and the journals Genetics and G3: Genes, Genomes, Genetics, have opened up a new pipeline not only for data type flagging and entity curation but also for more interactive publishing that directly links entities in published papers to their respective WormBase Web pages. Such collaborations serve to more tightly couple the biomedical literature with online resources that curate and integrate their content.
Text-mining applications have helped tremendously with automating WormBase workflow, but additional data types might still benefit from text mining, and further improvements can be made to the precision and recall of existing methods. For example, although we, currently, use Textpresso and HMMs for GO CC and MF annotation, we have not yet employed text mining for GO BP annotation. Text mining might also help us to develop more sophisticated triage approaches that consider existing curated information when flagging a paper. Such an approach would allow curators to prioritize curation of truly novel experimental results.
Textpresso for cellular component curation at dictyBase and TAIR
As part of the BioCreative workshop, we also evaluated the performance of a Textpresso for CCC system for dictyBase. Subsequent to the workshop, we performed a similar evaluation for TAIR. Results of the evaluation indicate that, similar to the C. elegans results (7), the use of a Textpresso-based annotation system for CCC results in annotations of high precision for both dictyBase and TAIR. This indicates that when curators are presented with Textpresso sentences, they are usually able to make the correct annotation of appropriate granularity.
In all cases, however, annotation recall is lower than precision. As with C. elegans, lower annotation recall appears to be due to a combination of factors, including failed gene product recognition, description of some experimental results over several sentences, incompleteness of Textpresso categories and true-positive statements that report experimental results without using terms from each of the required search categories.
In some of these cases, recall could be improved through fairly straightforward steps, such as adding new terms to the Textpresso categories and improving gene product recognition. For example, dictyBase contains distinct gene products referred to as myosin and myosin IE. The current implementation of Textpresso for dictyBase recognized myosin correctly, but not the phrase ‘myosin IE’. Thus, any annotations to myosin IE in the evaluation set could not be made. Further, gene or protein names containing Greek characters were not recognized; solutions to this problem will need to be handled at the level of document conversion (PDF-to-text).
Although additional terms can always be added to the appropriate Textpresso categories, alternative or complementary search strategies may prove to be equally useful. For example, allowing curators to view sentences that contained matches to less than the four categories typically used for searches might help identify additional annotations that can be made. This strategy might be especially productive if such sentences were ranked according to the presence of terms or phrases with the highest probability of describing an experimental result, such as GFP, signal or co-localized. Further analysis will need to be performed to determine the utility of such an approach.
As part of the Textpresso evaluation, we also examined the effect of including a fifth Textpresso category that includes terms such as ‘Figure’ and ‘Fig’ on performance metrics. We were interested to learn whether a more restrictive search that only returned sentences referring to a figure in the paper was sufficient for curation and might result in greater search precision. Our analyses suggest that although including this fifth category can improve the precision of sentences returned, it can also lower recall. Further, including this fifth category did not appear to have dramatic effects on annotation precision and in fact slightly lowered annotation recall. As a result, we will likely continue to perform the less restrictive four-category searches. Further improvements to precision might instead include a document-filtering step based on SVM or HMM document classification, as well as a filtering step that restricts searches to particular sections of papers. Currently, as WormBase uses an SVM for classifying documents with respect to expression pattern data and as Textpresso has the ability to search specific paper sections, at least two of these options could readily be implemented for the dictyBase and TAIR curation pipelines.
Further improvements to the Textpresso for cellular component curation pipeline
In addition to category updates, filtering steps and technical improvements, analysis of the curation workflow provides further insight into how this curation pipeline may be improved. Currently, the Textpresso for CCC tool presents sentences in isolation from the rest of the paper, i.e. without surrounding context. Curators are used to evaluating information in the context of the whole paper, however, so an additional improvement to Textpresso-based pipelines would be presentation of search results in the context of the full article, with the flexibility to view only those sentences that reside within sections of a paper most likely to contain curatable information, i.e. Results and Figure Legends. Such a display would more closely mimic the actual environment in which curators make annotations and eliminate the need for curators to open up a separate Web page to cross-check Textpresso sentences with the full text of the paper. Also, as the Textpresso system currently requires acquisition and storage of full-text documents, ‘on-the-fly’ processing may be particularly useful for open-access articles and would allow curators to assess the content of a paper without having to download and store the document.
The initial CCC curation tool was developed to complement an existing GO curation tool at WormBase and was designed to capture a very specific type of annotation, namely GO CC annotations that could be made using the Inferred from Direct Assay evidence code. As use of this tool is increasing, however, we hope to expand its functionality by allowing for additional types of annotation, including annotations using the GO annotation qualifier ‘NOT’ and annotations made using the Inferred from Physical Interaction evidence code. We would also like to make the curation tool more interactive, allowing for users to more easily correct or change annotations and add new gene names or Textpresso category terms while annotating. The latter feature would allow users to leverage their curation efforts to improve the system as well as expand their database’s catalogue of gene names and synonyms. Similarly, providing feedback from the curation form to the GO would help improve the representation of subcellular organelles, and especially their published synonyms, in the CC ontology.
We found that Textpresso can be used by both dictyBase and TAIR to more efficiently curate GO CC annotations. To what extent could Textpresso be used to improve curation efficiency of the literature of other organisms such as human or mouse that have even larger bodies of literature and where precise species identification can be difficult? Handling of large corpora is not problematic, as Textpresso can distribute huge corpora among several machines and then use Web services to query the subdivided corpora, collating and analyzing the results on a master node.
For mammalian species, however, paper identification for curation is not always straightforward, and thus, subsequent text-mining approaches can be potentially more challenging. In this context, Textpresso may need to be used in a different manner, one that perhaps emphasizes triage based on experimental results first and precise species identification second. For the latter task, Textpresso categories that specifically search for sentences describing how an experiment was performed (e.g. what cell lines, DNA constructs or siRNA targets) might be especially valuable. Textpresso for Mouse (http://www.textpresso.org/mouse/) is one of the organism-specific sites currently in production, allowing us to begin exploring effective ways to use Textpresso in mammalian curation pipelines.
In summary, continued use of, and familiarity with, the Textpresso system by curators combined with procedural improvements could readily provide greater increases to curation efficiency over time. Expanding a Textpresso-based curation strategy to a broader range of data types and organisms might thus help to improve curation efficiency overall, speeding up the rate at which new biological knowledge is incorporated into, and made useful by, model organism databases.
Funding
The US National Human Genome Research Institute (HG02223 to WormBase) and the British Medical Research Council (G070119 to WormBase); The US National Human Genome Research Institute (HG004090 to Textpresso); The US National Institute of Health (GM64426 to dictyBase); The National Science Foundation (DBI-0850219 to TAIR], with additional support from TAIR sponsors (http://www.arabidopsis.org/doc/about/tair_sponsors/413); The National Science Foundation (0822201 to The Plant Ontology); US National Human Genome Research Institute (HG002273 to The Gene Ontology Consortium). PWS is an investigator with the Howard Hughes Medical Institute. Funding for open access charge: US National Human Genome Research Institute [Grant no. HG002273].
Conflict of interest. None declared.
Acknowledgements
We would like to thank the BioCreative Workshop 2012 Steering Committee for the opportunity to participate in the workshop and, in particular, C. Arighi for advice and support regarding the Task III evaluation. We also thank C Grove, K Howe, R Kishore, D Raciti, MA Tuli, X Wang, G Williams and K Yook for their helpful comments on the manuscript and gratefully acknowledge S. Wimpfheimer for assistance with the figures. The members of the WormBase Consortium are M. Berriman and R. Durbin (Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK); T. Bieri, P. Ozersky and J. Spieth (The Genome Institute, Washington University School of Medicine, St. Louis, MO 63108, USA); A. Cabunoc, A. Duong, T.W. Harris and L. Stein (Ontario Institute for Cancer Research, 101 College Street, Suite 800, Toronto, ON, Canada M5G0A); J. Chan, W.J. Chen, J. Done, C. Grove, R. Kishore, R. Lee, Y. Li, H.M. Muller, C. Nakamura, D. Raciti, G. Schindelman, K. Van Auken, D. Wang, X. Wang, K. Yook and P.W. Sternberg (Division of Biology, California Institute of Technology, 1200 E. California Boulevard, Pasadena, CA 91125, USA); J. Hodgkin (Genetics Unit, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, United Kingdom); P. Davis, K. Howe, M. Paulini, M.A. Tuli, G. Williams and P. Kersey (EMBL-European Bioinformatics Institute Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK).