-
PDF
- Split View
-
Views
-
Cite
Cite
Donghui Li, Tanya Z. Berardini, Robert J. Muller, Eva Huala, Building an efficient curation workflow for the Arabidopsis literature corpus, Database, Volume 2012, 2012, bas047, https://doi.org/10.1093/database/bas047
Close - Share Icon Share
Abstract
TAIR (The Arabidopsis Information Resource) is the model organism database (MOD) for Arabidopsis thaliana, a model plant with a literature corpus of about 39 000 articles in PubMed, with over 4300 new articles added in 2011. We have developed a literature curation workflow incorporating both automated and manual elements to cope with this flood of new research articles. The current workflow can be divided into two phases: article selection and curation. Structured controlled vocabularies, such as the Gene Ontology and Plant Ontology are used to capture free text information in the literature as succinct ontology-based annotations suitable for the application of computational analysis methods. We also describe our curation platform and the use of text mining tools in our workflow.
Database URL:www.arabidopsis.org
Introduction
Published literature continues to be one of the most important repositories of scientific data. The number of articles in PubMed related to Arabidopsis thaliana, a model organism for plant biology research, has increased from 2014 articles in 2002 to 4343 in 2011. Accessing the huge volume of experimental results in the primary research literature, often published in the form of unstructured free text, poses a significant challenge for the research community. To meet this challenge, the biocuration community has taken on the task of collecting and organizing published experimental data into a format suitable for large-scale querying, comparison and computational analysis (1). This is achieved by converting some of the free text data into controlled vocabulary-based statements through manual curation of the primary literature. Biologists today have become increasingly dependent on such computable datasets provided by biological databases for data access, analysis and discovery.
TAIR (The Arabidopsis Information Resource, http://www.arabidopsis.org) is the primary database for A. thaliana (2, 3). TAIR serves as a centralized gateway to Arabidopsis biology, research materials and community members. TAIR is highly used by Arabidopsis researchers, as well as the broader plant research community, with Google Analytics usage statistics showing 164 000 visits and 53 000 unique visitors per month on average over the past year. Data available from TAIR includes the complete A. thaliana genome sequence along with gene structures, gene function information, metabolic pathways, gene expression patterns, genome maps, genetic and physical markers, publications, biological materials including seed and DNA stocks and information about the Arabidopsis research community. TAIR is a manually curated database; data are processed by trained biocurators to ensure that annotations accurately capture experimental results. TAIR data come from a variety of sources, including curator review of published literature, in-house and external computational pipelines for annotating gene structure and function, integration of data from other biological databases and resources [GenBank, ABRC (Arabidopsis Biological Resource Center), UniProt, Gene Ontology Consortium, etc.] and submissions from the research community, ranging from data on a single gene to large genomics, transcriptomics and proteomics datasets. TAIR also provides researchers with an extensive set of data visualization and analysis tools.
Manual literature curation has been an important task for TAIR curators since the database’s inception in 1999. This labor-intensive process produces consistent and high-quality annotations. However, an important challenge for us, as well as curators at many other biological databases, is finding a way to handle the ever-increasing number of publications with limited curation resources. We employ a three-pronged approach to address this challenge: first, we have developed a streamlined curation workflow incorporating both automated and manual elements to cope with the flood of new research articles. Second, we use text mining methods for entity recognition and extraction to speed up curation; to this end, we have developed and improved PubSearch (4), an integrated curation platform capable of article prioritization, gene recognition and the annotation of multiple entities (gene function, expression, polymorphism, phenotype, etc.) from full-text literature using controlled vocabularies or free text. Recently, we have also developed a semi-automated curation system for protein subcellular localization using Textpresso in collaboration with the WormBase team at Caltech (http://www.wormbase.org). Details of this project are presented in an accompanying article (Van Auken, K. Wei, C.H. et al., submitted for publication). Third, to complement our in-house curation effort, we developed a novel journal collaboration program to collect annotations directly from authors at the time of publication (5, 6).
Here, we describe TAIR’s literature curation workflow as presented at the BioCreative (Critical Assessment of Information Extraction Systems in Biology) 2012 Workshop (http://www.biocreative.org). We briefly describe the PubSearch curation platform and highlight the use of text mining in our curation workflow. One of the major objectives of the BioCreative project is to apply text mining to biocuration (7, 8). We provide a detailed description of TAIR’s curation workflow to enable discovery of bottlenecks that can be addressed by additional text mining solutions.
Scope of literature curation at TAIR
We aim to extract comprehensive Arabidopsis gene-related information from published literature. With that goal in mind, we capture the following data: gene function (e.g. molecular function, biological process, subcellular localization); gene expression pattern (including anatomical parts and developmental stages where the gene is expressed); alleles; phenotypes; gene symbols and full gene names. We no longer curate protein–protein interactions from the literature, because we can import these from other resources dedicated to this task, e.g. BioGRID (9).
Information extracted from the literature is integrated into the TAIR database, which serves as a central access point for Arabidopsis data. For each gene, we maintain a regularly updated page, the Locus Detail page (e.g. http://www.arabidopsis.org/servlets/TairObject?name=AT3G52910&type=locus), which summarizes the current set of relevant information present in TAIR.
As of 31 August 2012, we have collected 41 608 Arabidopsis research article records published between 1947 and 2012. Of these, 26 218 (63%) are tagged as potentially containing gene-related information based on the mention of an Arabidopsis gene name in the abstract or full text. Within this subset, 8820 articles (34%) have been used to make controlled vocabulary annotations.
The Arabidopsis genome contains 28 775 genes encoding protein or RNA products (3). Of these, 25 962 (90%) now have at least one publication associated to them within TAIR. About 18 797 (65.3%) genes are associated to articles which have 100 or fewer genes linked to them. About 10 801 (37.5%) genes are associated to articles which have 10 or fewer genes linked to them. The number of genes with at least one Gene Ontology (GO) annotation or Plant Ontology (PO) annotation (including both experimental and non-experimental evidence codes) from an article has reached 16 898 (59%) and 20 012 (70%), respectively. The number of genes with at least one experimental GO annotation or PO annotation from an article is 11 806 (41%) and 20 009 (70%), respectively. We carried out a pilot study in 2011 to investigate whether the set of genes lacking any GO or PO annotation but having an associated paper could be annotated from data in those papers. For the 289 genes in the pilot study, most of the associated papers placed the gene within a family based on sequence similarity but provided no functional characterization information. We additionally searched for papers relevant to this set of genes using Textpresso for Arabidopsis and Google Scholar (scholar.google.com) but were not able to retrieve any additional references that could be used to annotate gene function.
Curation tools
We have developed a web-based literature curation tool, PubSearch, that serves as the main curation platform for TAIR (4). PubSearch consists of two components: one component handles article ranking and curation task management among curators. The other component is a web-based literature curation tool that handles article retrieval and gene name recognition and allows curators to make annotations or add free text descriptions to capture the research results found in the literature. It is based on a MySQL relational database accessed through Java Servlet and Java Server Pages running in a Tomcat container for the API and front-end applications. We have made extensive improvements to this tool since its initial development, adding new features such as community annotation processing capability and database-driven full-text downloading. Data entered into the PubSearch curation database are propagated to the TAIR production database for public use. The data transfer is supported by extract–transform–load (ETL) pipelines that move data between the PubSearch and TAIR databases on a regular basis.
Curation workflow
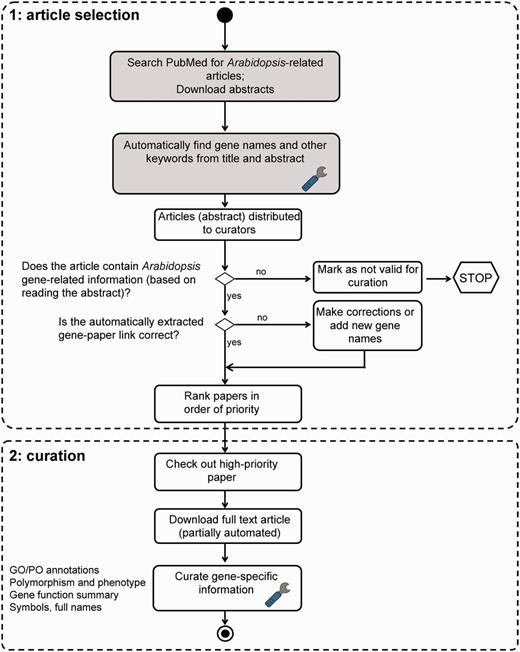
Figure 1 illustrates our manual literature curation workflow. The operation can be divided into two phases: (i) article selection and (ii) curation. Selection of articles for curation (phase A) is based on abstracts only; full-text articles are used for curation (phase B).
TAIR literature curation workflow. The curation process consists of both automated (shaded boxes) and manual steps. Full text PDF download is partially automated. PubSearch automatically downloads PDFs from selected journals, other articles not retrieved by PubSearch are manually downloaded by curators. Potentially curatable papers are identified by searching PubMed for ‘Arabidopsis’. Gene names are automatically identified from abstracts, then manually verified by curators. Articles that contain Arabidopsis gene-related information are flagged by curators for manual curation. Paper ranking is based on a combination of journal impact factor and manual prioritization. Papers describing previously uncharacterized genes are given the highest priority. Curators then proceed to extract comprehensive gene-specific information from high-priority papers. Steps that involve text mining are indicated by the wrench icon.
Phase A. Article selection
The selection of articles for full curation is crucial for the efficient use of scarce curator resources. We employ a multi-step process to progressively filter out articles less likely to contain information that can be integrated into the current TAIR database structure. At the end of the article selection process the remaining papers are highly likely to contain new information that can be captured with the curation tools and practices used at our MOD.
Article identification and retrieval
We download and prioritize all incoming new articles before moving on to curation. At the beginning of each month, we query for and download all article records from PubMed that contain ‘Arabidopsis’ in the title, abstract or keywords. This article identification and retrieval step is performed by the PubSearch literature curation tool. PubSearch uses the NCBI (National Center for Biotechnology Information) Entrez Utilities web services generated for Java (10) to access the PubMed library, search for articles and download the publication details, including the abstract.
Gene name recognition
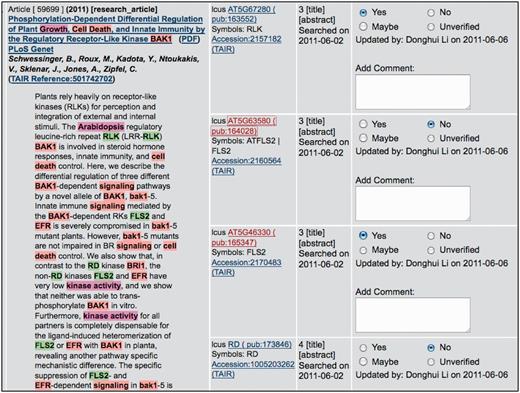
Gene names and other keywords are automatically identified within the downloaded titles and abstracts, using an extensive set of ontology terms, gene names and gene symbols stored within the PubSearch database. PubSearch uses an Aho-Corasick search-tree algorithm to search article titles and abstracts for these keywords (11). Each match is stored as a prospective hit, an association between a keyword and the article, in the PubSearch database (Figure 2). A small subset of valid Arabidopsis gene symbols are intentionally ignored in the hit generation process. This subset includes gene symbols identical to common English words, prefixes or abbreviations, such as FOR, CO, AND and LTD, that appear so frequently in article abstracts that manually reviewing each hit becomes unacceptably time-consuming. For gene names falling within this subset, association of articles to genes is performed manually. We have not developed an automated mechanism for detecting occurrences of these names that are likely to be real gene mentions; the number of gene names in this set is small and development of such a mechanism would not be expected to have a significant effect on our productivity.
PubSearch gene name recognition interface. The left panel displays article abstract with various entities (gene name: e.g. FLS2, species name: e.g. Arabidopsis and keywords: e.g. kinase activity) highlighted. The highlighted entities are automatically identified by PubSearch. Candidate gene identifiers suggested by PubSearch are displayed in center panel. The right panel allows curators to record the results of manual verification. In this example, the symbol FLS2 in the abstract is mapped to two gene identifiers: AT5G63580 and AT5G46330. Curators link FLS2 to the correct gene identifier by analyzing the context in the abstract, as well as existing annotations to these two genes. Due to space constraint, this figure only displays a partial list of candidate gene identifiers suggested by PubSearch. For example, BAK1, which corresponds to AT4G33430 is not shown in this figure.
A manual verification step follows the initial automated gene name recognition process. This is necessary because some gene symbols occur redundantly and this ambiguity cannot be automatically resolved by PubSearch. For example, the symbol FLS2 is used in the literature to describe two different A. thaliana genes, AT5G46330 (FLAGELLIN-SENSITIVE 2) and AT5G63580 (FLAVONOL SYNTHASE 2), encoding a leucine-rich repeat serine/threonine protein kinase and a flavonol synthase, respectively. When FLS2 appears in an abstract, PubSearch creates two hits for this symbol, to AT5G46330 and AT5G63580. In the manual validation step, a curator verifies the match to the correct gene and invalidates the other match based on the context in the abstract and gene-related information including full names and previous annotations. In most cases, gene disambiguation can be achieved by reading the abstract. However, in some cases, curators must go on to read the full text and/or perform an analysis (e.g. BLAST search of sequences reported in the article) in order to disambiguate gene symbols. In such cases, this task is deferred to the curation phase.
In addition to gene name and symbol recognition, PubSearch includes a script that identifies newly assigned A. thaliana gene names and adds them to the database. This script searches titles and abstracts using the regular expression ‘\b(?:At)?[A-Z]{2,4}\d{1,3}\b’. This regular expression is designed to identify symbols containing two-to-four fully capitalized letters followed by one-to-three digits, e.g. FLS5, with or without the prefix ‘At’ representing the species A. thaliana. About half of putative new gene symbols discovered in this way are true positives. The remaining candidates are most often either valid gene symbols from other species or names of chemicals, strains or restriction sites. False positives resulting from this process are manually removed by curators.
Article priority ranking
Articles are prioritized using a combination of automated ranking based on journal impact factor and curator manual prioritization, allowing us the flexibility to adjust rankings to reflect shifting curation resources and priorities. A priority ranking (high, medium, normal) is automatically assigned to each article based on journal-impact factor. Following this step, curators review all new abstracts to determine whether the article contains Arabidopsis gene-related information. If the answer is ‘No’, the article is not curated but still remains part of the literature corpus that can be accessed through TAIR. For articles that do contain Arabidopsis gene-related information, curators manually verify the gene–article links as described above and make corrections when necessary. Regardless of the initial rank based on journal impact factor, if the article describes characterization of a novel gene, the priority is changed by the curator to ‘first’, the highest priority setting.
The flexibility of this combined approach to article ranking has allowed us to maximize the impact of our limited curation resources. Prior to 2010, with a larger curation team, we were able to curate all articles published in high-impact journals (first and high priority papers). In the past 2 years, due to decreased funding for manual literature curation, we have limited our curation efforts mainly to ‘first priority’ articles describing the characterization of previously undescribed (‘novel’) genes. This approach maximizes the number of Arabidopsis genes with at least one experimentally based functional annotation, at the expense of adding additional annotations to genes for which some information is already known.
Phase B. Curation
To enable curation of full text, PubSearch scripts download PDF versions of articles from selected journals licensed to Stanford University. This is accomplished using a database-driven system that periodically accesses publishers’ web sites, downloading full-text versions of Arabidopsis articles obtained from our PubMed search. We store these copyrighted materials securely and tightly control access to them through firewalls and application-level security settings.
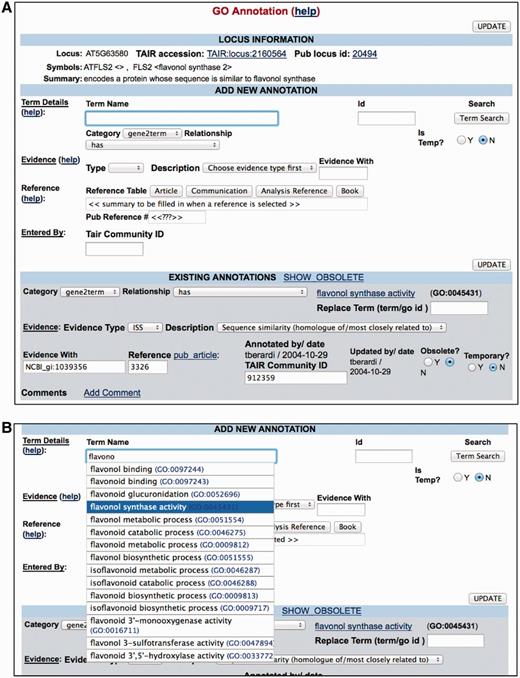
To begin the curation process, a curator selects an article for curation and moves it from the general queue to a personal article list to prevent simultaneous curation by other curators. The article is read and any relevant results are extracted and entered into the PubSearch curation interface, as shown in Figure 3. Upon completion of data entry, the curator marks the article as scanned (i.e. curated) and enters the date of curation.
PubSearch GO annotation interface. (A) Top panel Locus Information provides a summary of gene function. The ‘Add New Annotation’ section allows curators to compose all essential components required for a GO annotation. Lower panel displays existing annotations. (B) The built-in GO term auto-complete functionality allows curators to select from a list of GO terms as they type. The same interface is also used for PO annotations.
Use of ontologies and controlled vocabularies
We make extensive use of the GO (12) and PO (13) structured controlled vocabularies to convert the free text information in the literature into annotations in a standardized format. The resulting annotations are entered into a relational database, enabling querying and computational analysis. Each annotation includes a reference that connects the annotation to the original information source. TAIR curators follow the best practice guidelines developed by the Gene Ontology Consortium and the wider biocuration community.
Table 1 summarizes data elements curated in the TAIR workflow and the controlled vocabularies used. The Arabidopsis community has developed a nomenclature system in which each gene is assigned a unique AGI (Arabidopsis Genome Initiative) locus identifier in a standardized format (e.g. AT5G46330). TAIR is currently the central agency responsible for assigning Arabidopsis locus identifiers. Many genes also have other names in the literature (e.g. AT5G46330 is commonly known as FLAGELLIN-SENSITIVE 2 or FLS2). We maintain an extensive list of gene names that includes the AGI code, gene symbol, gene full name and reference where the gene symbol was found (e.g. AT5G46330-FLS2- FLAGELLIN-SENSITIVE 2-reference 1, or AT5G63580-FLS2- FLAVONOL SYNTHASE 2-reference 2). During the curation process, we manually verify that the gene symbol and full name reported in the article are present in the gene name list and are mapped to the correct AGI locus identifier.
Data elements extracted from literature and controlled vocabularies used in curation
| Data elements in literature . | Controlled vocabularies . | Examples and links to CV (if available) . |
|---|---|---|
| Gene | TAIR locus identifiers | AT5G46330 |
| Gene function | GO | GO:0009908 flower development http://amigo.geneontology. org/cgi-bin/amigo/browse.cgi |
| Gene expression pattern | PO | PO:0009046 flower http://www.plantontology.org/ |
| Polymorphism | TAIR-controlled vocabulary | Insertion, substitution |
| Phenotype | Free text transitioning to plant-specific Phenotype Ontology | Female sterile; altered ovule development; integuments fail to cover nucellus; reduced plant height; reduced length in inflorescence internodes; reduced levels of pollen, smaller leaves; late flowering, slow growth. |
| Data elements in literature . | Controlled vocabularies . | Examples and links to CV (if available) . |
|---|---|---|
| Gene | TAIR locus identifiers | AT5G46330 |
| Gene function | GO | GO:0009908 flower development http://amigo.geneontology. org/cgi-bin/amigo/browse.cgi |
| Gene expression pattern | PO | PO:0009046 flower http://www.plantontology.org/ |
| Polymorphism | TAIR-controlled vocabulary | Insertion, substitution |
| Phenotype | Free text transitioning to plant-specific Phenotype Ontology | Female sterile; altered ovule development; integuments fail to cover nucellus; reduced plant height; reduced length in inflorescence internodes; reduced levels of pollen, smaller leaves; late flowering, slow growth. |
CV, controlled vocabularies
Data elements extracted from literature and controlled vocabularies used in curation
| Data elements in literature . | Controlled vocabularies . | Examples and links to CV (if available) . |
|---|---|---|
| Gene | TAIR locus identifiers | AT5G46330 |
| Gene function | GO | GO:0009908 flower development http://amigo.geneontology. org/cgi-bin/amigo/browse.cgi |
| Gene expression pattern | PO | PO:0009046 flower http://www.plantontology.org/ |
| Polymorphism | TAIR-controlled vocabulary | Insertion, substitution |
| Phenotype | Free text transitioning to plant-specific Phenotype Ontology | Female sterile; altered ovule development; integuments fail to cover nucellus; reduced plant height; reduced length in inflorescence internodes; reduced levels of pollen, smaller leaves; late flowering, slow growth. |
| Data elements in literature . | Controlled vocabularies . | Examples and links to CV (if available) . |
|---|---|---|
| Gene | TAIR locus identifiers | AT5G46330 |
| Gene function | GO | GO:0009908 flower development http://amigo.geneontology. org/cgi-bin/amigo/browse.cgi |
| Gene expression pattern | PO | PO:0009046 flower http://www.plantontology.org/ |
| Polymorphism | TAIR-controlled vocabulary | Insertion, substitution |
| Phenotype | Free text transitioning to plant-specific Phenotype Ontology | Female sterile; altered ovule development; integuments fail to cover nucellus; reduced plant height; reduced length in inflorescence internodes; reduced levels of pollen, smaller leaves; late flowering, slow growth. |
CV, controlled vocabularies
Gene function data presented in the literature are captured using GO vocabularies consisting of molecular function, biological process and cellular component terms. We use PO vocabularies for plant anatomy and plant growth and developmental stages to annotate gene expression. GO evidence codes and references are attached to each annotation to provide provenance, as well as a pointer to the original published result. Examples of GO and PO annotations are shown in Table 2.
Examples of GO and PO annotation in TAIR
| . | Gene . | Relationship type . | Term . | Evidence type . | Reference . | Annotated by and date . |
|---|---|---|---|---|---|---|
| GO annotation | AT5G46330 | involved in | defense response to bacterium GO:0042742 | inferred from mutant phenotype: analysis of physiological response | Xiang et al. (2007) | The Arabidopsis Information Resource/ 2008-01-15 |
| PO annotation | AT5G46330 | expressed in | root tip PO:0000025 | inferred from direct assay: localization of GFP/YFP fusion protein | Robatzek et al. (2006) | The Arabidopsis Information Resource/ 2007-04-04 |
| . | Gene . | Relationship type . | Term . | Evidence type . | Reference . | Annotated by and date . |
|---|---|---|---|---|---|---|
| GO annotation | AT5G46330 | involved in | defense response to bacterium GO:0042742 | inferred from mutant phenotype: analysis of physiological response | Xiang et al. (2007) | The Arabidopsis Information Resource/ 2008-01-15 |
| PO annotation | AT5G46330 | expressed in | root tip PO:0000025 | inferred from direct assay: localization of GFP/YFP fusion protein | Robatzek et al. (2006) | The Arabidopsis Information Resource/ 2007-04-04 |
Adapted from TAIR locus detail page:
Examples of GO and PO annotation in TAIR
| . | Gene . | Relationship type . | Term . | Evidence type . | Reference . | Annotated by and date . |
|---|---|---|---|---|---|---|
| GO annotation | AT5G46330 | involved in | defense response to bacterium GO:0042742 | inferred from mutant phenotype: analysis of physiological response | Xiang et al. (2007) | The Arabidopsis Information Resource/ 2008-01-15 |
| PO annotation | AT5G46330 | expressed in | root tip PO:0000025 | inferred from direct assay: localization of GFP/YFP fusion protein | Robatzek et al. (2006) | The Arabidopsis Information Resource/ 2007-04-04 |
| . | Gene . | Relationship type . | Term . | Evidence type . | Reference . | Annotated by and date . |
|---|---|---|---|---|---|---|
| GO annotation | AT5G46330 | involved in | defense response to bacterium GO:0042742 | inferred from mutant phenotype: analysis of physiological response | Xiang et al. (2007) | The Arabidopsis Information Resource/ 2008-01-15 |
| PO annotation | AT5G46330 | expressed in | root tip PO:0000025 | inferred from direct assay: localization of GFP/YFP fusion protein | Robatzek et al. (2006) | The Arabidopsis Information Resource/ 2007-04-04 |
Adapted from TAIR locus detail page:
We have developed sets of controlled vocabulary terms to capture polymorphism information (e.g. polymorphism type, mutagen) from the literature. At the same time, we allow a curator-created free text description to be attached to a polymorphism. Germplasm refers to a strain with a unique genotype. We have also developed sets of controlled vocabulary terms to capture germplasm information such as species variant (typically a lab strain or natural variant, known as an ecotype in the Arabidopsis community), alleles known to be present in the germplasm, etc. We also allow a free text description to be attached to a germplasm. Phenotypes are currently described using free text. We are working with the community to supplement these phenotype descriptions with ontology-based phenotype annotations using PO, GO, ChEBI (Chemical Entities of Biological Interest) (14, 15) and PATO (Phenotypic Quality Ontology) (16). As in the case of the GO and PO annotations, references are provided for the phenotypes to allow the users to refer back to the original published result. Table 3 shows how polymorphism and phenotype data are represented within TAIR.
Representation of polymorphism and phenotype in TAIR
| Polymorphism name . | Gene ID . | Polymorphism type . | Polymorphism site . | Inheritance . | Germplasm . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|---|
| fls2-17 | AT5G46330.1 | substitution | exon | recessive | FLS2-17 | Mutant seedlings treated with 10-μM flg22 peptide (strong growth inhibitor) display shoot and root growth similar to that of wild-type Ler. | Gomez-Gomez et al., 2000 |
| Polymorphism name . | Gene ID . | Polymorphism type . | Polymorphism site . | Inheritance . | Germplasm . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|---|
| fls2-17 | AT5G46330.1 | substitution | exon | recessive | FLS2-17 | Mutant seedlings treated with 10-μM flg22 peptide (strong growth inhibitor) display shoot and root growth similar to that of wild-type Ler. | Gomez-Gomez et al., 2000 |
Adapted from TAIR polymorphism page:
Representation of polymorphism and phenotype in TAIR
| Polymorphism name . | Gene ID . | Polymorphism type . | Polymorphism site . | Inheritance . | Germplasm . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|---|
| fls2-17 | AT5G46330.1 | substitution | exon | recessive | FLS2-17 | Mutant seedlings treated with 10-μM flg22 peptide (strong growth inhibitor) display shoot and root growth similar to that of wild-type Ler. | Gomez-Gomez et al., 2000 |
| Polymorphism name . | Gene ID . | Polymorphism type . | Polymorphism site . | Inheritance . | Germplasm . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|---|
| fls2-17 | AT5G46330.1 | substitution | exon | recessive | FLS2-17 | Mutant seedlings treated with 10-μM flg22 peptide (strong growth inhibitor) display shoot and root growth similar to that of wild-type Ler. | Gomez-Gomez et al., 2000 |
Adapted from TAIR polymorphism page:
Use of text mining in curation
In our current literature curation workflow, we use an Aho–Corasick keyword-searching algorithm to extract gene names and other keywords from article titles and abstracts and create associations between the terms and the articles. A manual verification step follows to confirm that the gene–article link is valid. Manual extraction of gene names and other keywords from literature followed by adding the associations to the database is a tedious process; the use of this algorithm for generating the putative links therefore greatly improves efficiency.
During the curation process, curators frequently must consult additional articles, e.g. to disambiguate a gene symbol or to track down specific information such as the mutation sites in certain alleles. In collaboration with our team, the WormBase team at Caltech has applied the Textpresso text mining tool (17) to the TAIR Arabidopsis literature corpus to produce Textpresso for Arabidopsis (http://www.textpresso.org/arabidopsis/). This tool, housed at Caltech, is available to both TAIR curators and general users and allows users to search over 43 151 abstracts (including some conference abstracts) and 33 955 full-text publications related to Arabidopsis (numbers as of July 2012). Users can search using specific keyword categories including A. thaliana gene names, GO and PO terms or combinations of keywords to narrow their search results. Sentences that contain matching keywords are retrieved together with bibliographic information so that users can quickly confirm the usefulness of a particular article and link directly to the full text, if they have the appropriate subscriptions to the journals in question.
Recently, in collaboration with the same Textpresso team at Wormbase, we have developed a semi-automated curation process to identify articles with cellular component information and create annotations from them. In this approach (18), the entire available Arabidopsis full-text literature corpus is processed by Textpresso, sentences that contain Arabidopsis gene names, protein subcellular localization data, as well as assay-related words are extracted and GO annotations are suggested. A curator then manually validates each suggested annotation. Validated annotations are exported as a flat file consisting of required fields (gene identifiers, GO term identifiers, evidence codes, references, annotation date). A curator then reformats the file before loading it into TAIR. Details of this approach are reported in a separate publication (Van Auken, K. et al., submitted for publication).
Summary and future directions
Our workflow efficiently prioritizes a manageable set of articles for curation by a small and well-trained curation team. Though we currently focus on extracting data from recently published articles about genes that have not previously been characterized, our workflow is flexible enough to adapt to other prioritization schemes. The workflow can be easily reverted to the journal impact factor-based prioritization or adapted to a new priority, e.g. articles about a specific set of genes or a gene family. In the latter case, we would simply retrieve the set of papers associated with that specific gene set and mark them as ‘first’ priority.
Manual literature curation by professional curators produces accurate and consistent annotations. Yet, manual extraction of gene function information from the literature and conversion into ontology-based annotations is a labor-intensive process. On average, a curator spends about 4 min per abstract (Wei, C.H. et al., submitted for publication) for article selection (verification of gene mentions and article priority ranking). Time spent on full-text curation varies widely from 0.5 h to >4 h per article depending on the complexity of the paper. This very time-consuming, detail-oriented, manual process of interpreting the text to extract and convert gene functions into ontology-based annotations has been the major bottleneck in our current workflow. In recent years, TAIR’s curation team has been able to curate only a fraction of newly published Arabidopsis articles (∼30%). There is a strong incentive to develop a more effective text mining system to automate the extraction and conversion of gene function information from the research literature into annotations based on standard, community-developed ontologies of biological concepts. This translates into an urgent need for tools capable of publication retrieval, entity recognition (including gene name, gene function, expression pattern, polymorphism and phenotype) and mapping of free text descriptions of gene function to ontology terms, with the latter representing the most severe bottleneck in our workflow. The ongoing BioCreative workshops are demonstrating increasing sophistication in more straightforward tasks such as entity recognition (19), protein–protein interaction (20) and basic document triage. The BioCreative tasks relating to the user interface (21) and to ontology term mining are just getting underway and early results are very promising. Nevertheless, the holy grail, full automation of the literature curation process, is not visible on the horizon. To improve effectiveness and productivity in the curation process at TAIR in the relatively near future, it will be necessary to develop a fully integrated environment that assists a curator at all levels of the process described here. This vision requires four factors: first, a focus on incremental improvements that will assist the curator (rather than the more distant and possibly unreachable goal of fully automating the process); second, involving the scientific community in the curation process; third, providing tools that are easily integrated into common programing frameworks through web services or programing APIs and fourth, building frameworks and interfaces that use the latest human–computer interface theory and technology to improve community and curator productivity at the same time as introducing machine learning and automation.
Funding
The National Science Foundation (grant DBI-0850219); the National Institutes of Health National Human Genome Research Institute (NHGRI) (grant 5P41HG002273-09 for gene function curation, partial funding). Additional support for gene function curation comes from the TAIR sponsorship program (see http://arabidopsis.org/doc/about/tair_sponsors/413 for a complete list of sponsors). Funding for open access publication is provided by the National Science Foundation.
Conflict of interest. None declared.
Acknowledgements
We would like to thank the BioCreative 2012 Workshop Steering Committee for the opportunity to participate in the workshop.