-
PDF
- Split View
-
Views
-
Cite
Cite
Prudence Mutowo-Meullenet, Rachael P. Huntley, Emily C. Dimmer, Yasmin Alam-Faruque, Tony Sawford, Maria Jesus Martin, Claire O’Donovan, Rolf Apweiler, Use of Gene Ontology Annotation to understand the peroxisome proteome in humans, Database, Volume 2013, 2013, bas062, https://doi.org/10.1093/database/bas062
Close - Share Icon Share
Abstract
The Gene Ontology (GO) is the de facto standard for the functional description of gene products, providing a consistent, information-rich terminology applicable across species and information repositories. The UniProt Consortium uses both manual and automatic GO annotation approaches to curate UniProt Knowledgebase (UniProtKB) entries. The selection of a protein set prioritized for manual annotation has implications for the characteristics of the information provided to users working in a specific field or interested in particular pathways or processes. In this article, we describe an organelle-focused, manual curation initiative targeting proteins from the human peroxisome. We discuss the steps taken to define the peroxisome proteome and the challenges encountered in defining the boundaries of this protein set. We illustrate with the use of examples how GO annotations now capture cell and tissue type information and the advantages that such an annotation approach provides to users.
Database URL:http://www.ebi.ac.uk/GOA/ and http://www.uniprot.org
Introduction
With increasing amounts of biological information being published from a wide range of experimental initiatives, it has become necessary to make this information easily available to a range of investigators, particularly those working with large datasets within a systems biology setting. The Gene Ontology (GO) is a bioinformatics project developed by the Gene Ontology Consortium that aims to introduce consistency in the description of functional information pertaining to gene products (proteins or functional RNAs) (1). The GO consists of three ontologies used to describe the Biological Processes, Molecular Functions and Cellular Component attributes of a gene product. The GO is used to describe the normal molecular functions and biological processes that a gene product is involved in as well as capturing its localization in a normal/non-disease cell. Over 15 curation groups in the GO Consortium carry out manual and automatic annotations of gene products. UniProt is a central member of the Consortium whose curators review experimental evidence presented in peer-reviewed publications to provide detailed, high-quality descriptions of protein function (2). In addition, high-quality automatic GO annotations are also supplied to the UniProt GO annotation set by Ensembl, EnsemblGenomes, InterPro and UniProt annotation prediction pipelines. Such automatic pipelines differently exploit gene orthology data, protein sequence signatures and existing cross-references or keywords from external controlled vocabularies to infer that proteins have particular functions or subcellular locations (2, 3). The inclusion of high-quality, automatic annotation predictions ensures that the UniProt GO annotation dataset supplies maximally complete functional information to a wide range of proteins [>340 000 taxonomic groups (October 2012)] that is especially valuable for species with limited experimentally derived information where predicted annotations sometimes serve as the sole source of information.
Organelle-focused protein annotation—the human peroxisome
Peroxisomes are single membrane-bound organelles that are present in most eukaryotic cells and contain a variety of enzymes involved in numerous metabolic processes, including catabolism of fatty acids, D-amino acids, polyamines as well as the biosynthesis of plasmalogens and the pentose phosphate pathway (4). The need to understand better the function of peroxisomes has been driven mainly by the establishment of a link between this organelle and a variety of diseases closely linked with peroxisomal dysfunction (5), including neurological abnormalities. Several clinical diagnosis protocols for peroxisome-associated diseases have been developed that rely on quantifying peroxisomal enzyme activity or metabolite level (6). Peroxisomal diseases have been categorized as those caused by a single enzyme deficiency such as Refsum disease or diseases due to multiple enzyme/protein defects such as the Zellweger syndrome. We have chosen to analyse the function of all human proteins localized to the peroxisome in a bid to establish a definitive set of peroxisome proteins in human and, by analysing their annotations, obtain a better understanding of the biological knowledge currently available for this organelle. Databases such as PeroxisomeDB (7) and PeroxisomeKB (8) describe sets of peroxisomal proteins from different species, while ambitious, large-scale experimental projects to decipher the biological function of peroxisomes in health and disease (http://cordis.europa.eu/fetch?CALLER=OFFR_TM_EN&ACTION=D&RCN=9223) have recently led to a significant increase in the availability of peroxisomal experimental information, which makes this annotation project timely.
The aim of the manual curation initiative was to define, based on experimental information, the evidence available to support the set of known human peroxisomal proteins as well as to use the GO to capture the diverse functions carried out by proteins in this organelle.
Methods
Defining a human peroxisome protein set
The set of proteins with peroxisomal annotation (from both manual and predicted methods) was extracted from the reviewed (Swiss-Prot) section of the UniProtKB release 2011_11. The initial dataset included 126 UniProtKB/Swiss-Prot entries, which when compared with protein sets from other peroxisomal resources (PeroxisomeKB and PeroxisomeDB) was able to provide an inclusive set of known and uncharacterized peroxisomally located proteins.
Using this protein set, a comprehensive literature-based manual annotation drive was embarked on to capture all the experimental instances of peroxisomal subcellular location as well as recording the functional information for each protein. GO annotations were created with the appropriate evidence codes to inform the user of the type of supporting evidence that exists for making a particular functional statement (9). A total of 88 human proteins were identified as having peroxisomal localization based on experimental evidence from published literature.
The human peroxisome interactome and its functions
The IntAct protein–protein interaction database (10) was queried for high-quality binary interactions for the 88 human proteins that have experimental support for peroxisomal location (IntAct database release 154). The Cytoscape 2.8.2 network visualization software (www.cytoscape.org) (11) was used to show proteins annotated to ‘peroxisome; GO:0005777’ (or descendant terms) and their interacting partners.
A GO slim is a subset of terms that describe broad categories of processes/functions or subcellular locations that can provide an overview of the common attributes of a gene or gene product set (12). A GO slim grouping of annotations to proteins belonging to the human peroxisomal interactome was retrieved using the generic GO slim (http://www.geneontology.org/GO.slims.shtml). The Cytoscape plugin Mosaic version 1.0 (13) was used to retrieve biological process annotations for the interactome and to group proteins with related GO terms together. The plugin parameters selected were a node view range of 5–200 with node colour based on the GO biological process. The most granular term annotated to all proteins in a cluster was used as the cluster label.
Enrichment analysis comparison of yeast and human peroxisome proteins
A total of 64 Saccharomyces cerevisiae (NCBI taxonomy ID 4932) proteins with manual peroxisomal annotations were retrieved from the UniProt-GOA database on 4 August 2012 using the QuickGO web browser. An enrichment analysis of biological process annotations for both the human and yeast peroxisomal protein sets was carried out using ClueGO version 1.4 (14), an enrichment tool available as a plugin for Cytoscape. Annotations derived from predictive methods were excluded from these analyses. Comparison of enrichment results was carried out using the Advanced Network Merge network analysis tool option in Cytoscape. GO enrichment analysis was also conducted on the human peroxisomal protein set on annotations available on 22 June 2011. A separate GO enrichment was performed on the same protein set using ontology and annotations from 4 August 2012 representing the protein set after the manual annotation effort. At first start up to ClueGO, the plugin created a folder containing precompiled ontology files. An older version of the ontology was downloaded from the ontology archive (http://www.geneontology.org/ontology-archive/) and saved in the ClueGO plugin folder. The earlier version of the ontology selected enables a correct depiction of the GO enrichment analysis for the peroxisomal proteins before the focused annotation effort. This was necessary as the ontology is in a constant state of flux with terms added, obsoleted and modified continuously. Using the version of the ontology before the focused annotation provides a true reflection of the terms available for use in enrichment analysis before the focused annotation. Similarly, previous versions of the annotations are found on the GOA database ftp site (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/old/HUMAN/). For both enrichment analyses, the right-sided hypergeometric enrichment test was performed using ClueGO at a medium network specificity selection. The selected GO tree levels were a minimum of 3 and a maximum of 8 while each cluster was set to a minimum of between 2% and 5% genes. The GO term fusion setting was selected to minimize redundancy and the highest significance term enriched was used as the leading term for each cluster. Enrichment analysis for human and yeast comparison was carried out at GO tree levels 6–12 in order to identify more granular differences between the species.
Data availability
GO annotation data for a variety of species is available from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) and the Gene Ontology Consortium website. All manual and predicted annotation sets can be retrieved using the QuickGO and AmiGO web browsers. GO annotations in UniProtKB can be found in the ‘Ontologies’ section of an entry. GO annotation data updates in UniProtKB occur monthly with each release. GO annotation data are also cross-referenced by many other scientific databases.
Both the experimentally determined and predicted peroxisomal data sets described in this work are available for download from QuickGO (See Supplementary Files S1 and Supplementary Data). The yeast peroxisomal proteins list is available in Supplementary File S3. Term enrichment data are found in Supplementary File S4 and GO slim analysis files are also supplied in Supplementary File S5.
Results
Information captured in the manual annotation process
The manual annotation initiative for the human peroxisome proteins led to a total of 88 peroxisomal proteins obtaining full functional annotation from the currently available literature. Annotations to 296 non-peroxisomal proteins were also captured where functional data were determined alongside the peroxisomal proteins of interest, thus making full use of all papers manually curated. A total of 1551 annotations were created in this process.
Capturing negative experimental findings and literature conflicts
The curation approach employed in the GO Consortium encourages capture of all available, up-to-date experimental annotation whether presenting positive or negative function or localization statements. This allows the user to obtain a comprehensive overview of the data available for a particular protein and to assess overall support from the cited references.
The GO annotation format allows curators to capture negative statements about a protein’s role using the ‘NOT’ qualifier. This qualifier is used sparingly in GO annotation to capture negative experimental findings where other evidence may have predicted a positive involvement in the same role. The human peroxisomal protein GO annotation dataset includes negative data where peroxisomal localization was expected based on protein orthology or sequence similarity, but refuted based on published experimental findings. An example of this would be the MK protein (UniProtKB Q03426), which is not peroxisomal contrary to expectation (15).
Capturing the context of peroxisomal localization
For proteins whose current experimental localization support is conflicting, the GO annotation format assists capture of distinct, specifically referenced localization statements. The inclusion of PubMed references for annotations captured from published literature means that both positive and negative associations can be made based on the reference supplying the information. This provides the users with all available information and gives them an opportunity to review the evidence presented. An example is the MPV17 protein (UniProtKB P39210) with both positive and negative peroxisomal localization data.
Although peroxisomes are found in most eukaryotic cells, annotations capturing the peroxisomal localization of specific proteins (as shown in Table 1) can enable curators to rapidly link cell or tissue type information to protein function where such an association has been experimentally made. This will provide users with extra information pertaining to a localization annotation where such information is provided.
Capturing cell- and tissue-type information in a protein GO annotation using the annotation extension field
| Gene name . | GO terms . | Evidence . | Annotation extension . | PubMed identifier . |
|---|---|---|---|---|
| MAVS | GO:0005777 peroxisome | IDA | Part_of (CL:0000182) hepatocyte | 20451243 |
| (UniProtKB Q7Z434) | ||||
| EPHX2 | GO:0005777 peroxisome | IDA | Part_of (Uberon :0005151) metanephric proximal tubule | 16314446 |
| (UniProtKB P34913) | ||||
| POMC | GO:0005782 peroxisomal matrix | IDA | Part of (CL:0002559) hair follicle cell | 20810565 |
| (UniProtKB P01189) | ||||
| FAR1 | GO:0005777 peroxisome | IDA | Occurs in (CL:0000057) fibroblast | 20071337 |
| (UniProtKB Q8WVX9) |
| Gene name . | GO terms . | Evidence . | Annotation extension . | PubMed identifier . |
|---|---|---|---|---|
| MAVS | GO:0005777 peroxisome | IDA | Part_of (CL:0000182) hepatocyte | 20451243 |
| (UniProtKB Q7Z434) | ||||
| EPHX2 | GO:0005777 peroxisome | IDA | Part_of (Uberon :0005151) metanephric proximal tubule | 16314446 |
| (UniProtKB P34913) | ||||
| POMC | GO:0005782 peroxisomal matrix | IDA | Part of (CL:0002559) hair follicle cell | 20810565 |
| (UniProtKB P01189) | ||||
| FAR1 | GO:0005777 peroxisome | IDA | Occurs in (CL:0000057) fibroblast | 20071337 |
| (UniProtKB Q8WVX9) |
Capturing cell- and tissue-type information in a protein GO annotation using the annotation extension field
| Gene name . | GO terms . | Evidence . | Annotation extension . | PubMed identifier . |
|---|---|---|---|---|
| MAVS | GO:0005777 peroxisome | IDA | Part_of (CL:0000182) hepatocyte | 20451243 |
| (UniProtKB Q7Z434) | ||||
| EPHX2 | GO:0005777 peroxisome | IDA | Part_of (Uberon :0005151) metanephric proximal tubule | 16314446 |
| (UniProtKB P34913) | ||||
| POMC | GO:0005782 peroxisomal matrix | IDA | Part of (CL:0002559) hair follicle cell | 20810565 |
| (UniProtKB P01189) | ||||
| FAR1 | GO:0005777 peroxisome | IDA | Occurs in (CL:0000057) fibroblast | 20071337 |
| (UniProtKB Q8WVX9) |
| Gene name . | GO terms . | Evidence . | Annotation extension . | PubMed identifier . |
|---|---|---|---|---|
| MAVS | GO:0005777 peroxisome | IDA | Part_of (CL:0000182) hepatocyte | 20451243 |
| (UniProtKB Q7Z434) | ||||
| EPHX2 | GO:0005777 peroxisome | IDA | Part_of (Uberon :0005151) metanephric proximal tubule | 16314446 |
| (UniProtKB P34913) | ||||
| POMC | GO:0005782 peroxisomal matrix | IDA | Part of (CL:0002559) hair follicle cell | 20810565 |
| (UniProtKB P01189) | ||||
| FAR1 | GO:0005777 peroxisome | IDA | Occurs in (CL:0000057) fibroblast | 20071337 |
| (UniProtKB Q8WVX9) |
The ‘annotation extension’ field (16) of a GO annotation line enables the capture of such contextual information, including cell and tissue type, by cross-referencing ontologies such as the Cell Type (17) and Tissue Type (18).
Table 1 shows the association between a gene product identifier (UniProtKB accession) and a GO term with a reference supporting the association and the evidence derived from the reference. The annotation extension column (with contents in italics) shows how more detailed information can be added to an annotation to capture the full context of the experimental findings. Capturing tissue and cell type information as shown by the examples in Table 1 will allow users to benefit from the additional knowledge that, for instance, the peroxisomal localization for the MAVS protein (UniProtKB Q7Z434) was confirmed in the hepatocyte. Capturing of cell and tissue type information alongside subcellular localization and protein molecular function can enable users to quickly ascertain in which cells or tissues certain peroxisomal-based functions are performed. This added layer of information allows some comparison of peroxisomal abundance between different tissue and cell types of the body to be made. The caveat scholasticus for using this cell and tissue type information in human is that although useful, this information is less complete compared with other species due to issues regarding tissue availability.
Feature chain and isoform-specific peroxisomal localization
The GO annotation format allows curators to associate biological information to specific forms of a protein, so wherever the published evidence allows, the UniProt GO annotation set includes specific isoform and post-translationally modified protein data, using UniProtKB isoform and feature chain identifiers. This has allowed the detailed annotation of, for example, POMC (UniProtKB P01189); capturing the fact that in the pituitary gland, the proteins peptide chains beta-lipotropin and beta-endorphin localize to the peroxisomal matrix, whereas the corticotropin, melanotropin alpha and gamma are not peroxisomal. http://www.ebi.ac.uk/QuickGO/GProtein?ac=P01189 (19). An example of protein isoform annotation is the localization of the short isoform of ECI2 (UniProtKB O75521-2) to the peroxisome, whereas no evidence has as yet been reported of the same localization for the long isoform http://www.ebi.ac.uk/QuickGO/GProtein?ac=O75521(20). Isoform-specific localization information has been captured this way whenever appropriate evidence exists.
A biological process overview of the peroxisome interactome using GO
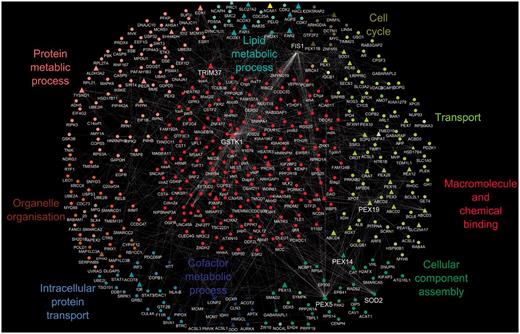
We constructed the peroxisome interactome as described in the ‘Methods’ section (The human peroxisome interactome and its functions). By combining publicly available interaction data for peroxisomal proteins with our GO annotation set, we are able to provide an overview of the biological processes of the peroxisome interactome. Figure 1 shows the peroxisome interactome overlaid with a GO slim view of the annotations that are associated with these proteins.
A protein–protein interaction map of the human peroxisome. The peroxisome proteins and their interacting partners comprised a set of 421 proteins with a total of 408 binary interactions; peroxisomal proteins are shown as triangles and non-peroxisomal proteins as circles. Protein–protein interactions are depicted as white edges. Multiple edges between two proteins represent interactions that have been identified by more than one approach. The generic GO slim was used to identify terms in the proteome. GO terms that were common to all proteins in a cluster are shown as the cluster label.
This analysis indicates that most peroxisomal proteins are involved in transport and co-factor or lipid metabolic processes, correlating with known peroxisomal functions. As peroxisomes do not have their own genetic material (21), transport plays a critical role in terms of protein import and export from the peroxisome as well as the transport of metabolites across the membrane. In addition, as 66% of human peroxisomal proteins are known to have an enzymatic function with metal ions as prerequisites for their activity, a high involvement in co-factor metabolism is not surprising, whereas peroxisomal involvement in lipid and protein metabolic processes has been well established (22).
Hub proteins of the peroxisome interactome
The interactome in Figure 1 displays several protein groupings that have a protein at their centre. These ‘hubs’ represent proteins that have a large number of interactions in the protein–protein interaction network. Hub proteins identified from the network include PEX5 (UniProtKB P50542), PEX14 (UniProtKB O75381), TRIM37 (UniProtKB O94972), PEX19 (UniProtKB P40855), GSTK1 (UniProtKB Q9Y2Q3), FIS1 (UniProtKB Q9Y3D6) and SOD2 (UniProtKB P04179). All these central proteins have experimental evidence available from the literature demonstrating their peroxisomal localization except for SOD2. Proteins that form hubs in interaction networks are suggested according to the ‘centrality-lethality rule’ (23) to be those proteins that are essential within a cell/organism for carrying out particular functions and can be used to indicate additional, core peroxisomal activities.
An example would be TRIM37 (UniProtKB O94972), an E3 ubiquitin-protein ligase, with dual localization reported in peroxisomes and the cytosol, which binds tumour necrosis factor receptors. In the case of the peroxisome, it has been proposed that ubiquitination of its membrane proteins may serve as a molecular signal for the degradation of the organelle. Peroxisomes have a very short life span, on average, a half-life of 2 days has been reported (24), indicating a tightly controlled organelle degradation mechanism. Another hub protein, FIS1 (UniProtKB Q9Y3D6), is known to be involved in both peroxisomal and mitochondrial fission, suggesting an active role in regulating organelle numbers.
Inferences of biological roles from predictive GO annotations
In the functional analysis of peroxisomal proteins, we identified some proteins with experimental evidence for peroxisomal localization but with no further experimental evidence pertaining to their biological process or molecular function. These proteins include MARF1 protein (UniProtKB Q9Y4F3), ABCD4 (UniProtKB O14678), ACOX3 (UniProtKB O15254) and isoform 2 of CNOT1 (UniProtKB A5YKK6-2). These proteins, once characterized further may hold the key to revealing even more clues about the various functions of the peroxisome. In the interim, however, predictive methods are used to create GO annotations to fill in the knowledge gap, while guiding experimental design to enable characterization of these proteins. MARF1 is predicted by InterPro2GO and UniProtKB keywords to be involved in female meiosis and oogenesis. ABCD4 is predicted to be ATP binding and involved in transmembrane transport due to the presence of certain InterPro domains within its protein sequence and its similarity to other sequences, whereas ACOX3 has predicted roles in lipid and small molecule metabolic processes from both sequence similarity and the presence of InterPro domains.
By looking at a protein’s position in the interaction network and using GO annotations, we can formulate suggestions for the functions of uncharacterized proteins. According to the ‘guilt by association’ premise (25), insight into the roles of a protein can be suggested from its interaction with a protein of known function. The uncharacterized protein most likely will be involved in similar biological processes to the characterized protein. The peroxisomal ABCD4 protein has not been extensively characterized, but in the interaction network in Figure 1, ABCD4’s interacting partners have annotations to child terms of GO:0006810 transport. This suggests the ABCD4 protein could possibly be involved in transport processes in the cell, which is also supported by predictions from the InterPro2GO and UniProt automated pipelines that predict this protein’s involvement in transport.
Impact of organelle-focused annotation on term enrichment analysis
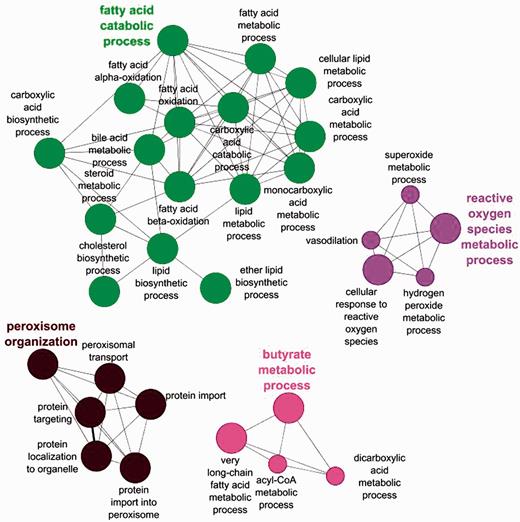
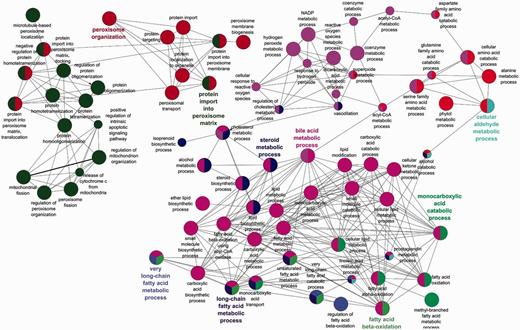
The benefits of an organelle-focused annotation approach can be demonstrated by performing the most commonly used type of GO analysis on this dataset: GO term enrichment. This method identifies terms that are over-represented within a given set of proteins compared with the whole proteome. We have compared the GO term enrichment of proteins with experimentally determined annotations obtained at the start of the initiative (Figure 2) with annotations currently available (Figure 3).
Biological process GO enrichment of 88 human peroxisome proteins before the focused manual peroxisome protein annotation effort. The circles represent enriched GO terms. The size of circle is proportional to the number of proteins containing the biological process term.
Biological process GO enrichment of 88 human peroxisome proteins after the focused manual peroxisome protein annotation effort. The circles represent enriched GO terms. The size of circle is proportional to the number of proteins containing the biological process term. Circles with different colours represent proteins that contain an intersection of GO terms.
Apart from the obvious increase in the number of enrichment terms in Figure 2 compared with Figure 3, new processes have also been enriched in the second analysis. Examples include ‘alcohol metabolic process and related terms’, ‘peroxisome fission’, ‘carboxylic acid biosynthetic process’ as well as ‘carboxylic acid catabolic process’ and ‘beta fatty acid oxidation’.
Added depth and specificity has also been added to the functional descriptions of the proteins. An example is the fatty acid metabolism role of the peroxisome, which due to the focused annotation resulted in more granular terms ‘fatty acid beta oxidation using acyl CoA oxidase’ being included in the annotation set thus giving better depth of information to the peroxisomal fatty acid oxidation process.
Focusing on an organelle for annotation also leads to growth of the ontology in that particular biological domain. In all, 49 new peroxisome-related terms were requested during the course of this project. The availability of these terms enables curators working in different groups to capture peroxisomal data in a consistent manner, which in turn facilitates systematic retrieval of information and data comparison.
Species comparison of peroxisomal proteomes: S. cerevisiae and human
As the human peroxisome proteome has now been comprehensively annotated, it was of interest to compare the functional attributes of the human peroxisome with those of the peroxisome from another species. One of the most well characterized and extensively annotated peroxisome proteomes is that of S. cerevisiae; there are currently 65 yeast proteins with experimentally determined peroxisomal subcellular localization (26). GO term enrichment was performed for the yeast peroxisome dataset and compared with that for the human peroxisome.
The differences observed in the S. cerevisiae and human peroxisome protein enrichment can be described by species differences, differences due to organelle function and a third category where the difference cannot be explained from the species or organelle point of view for reasons that will be discussed further.
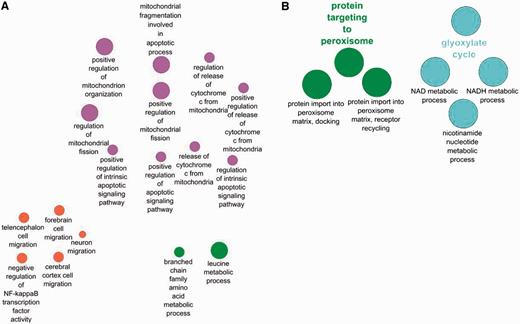
The GO term ‘glyoxylate cycle’ is seen only in the yeast enrichment and not in the human set. This is expected as the tricarboxylic acid cycle occurs in humans in the mitochondrion, not in the peroxisome. There are also a good number of terms that are enriched only in the human set and not in the yeast as a result of the differences in cell composition between the two species. Hence, terms like ‘forebrain cell migration’ and ‘neuron cell migrations’ are expected to be only found in the human set.
Terms enriched in the human set and depicted in Figure 4A highlight the importance of some human peroxisomal proteins in regulating mitochondrion organization and number as well as involvement in apoptosis. A similar enrichment was not observed for the yeast set. This may suggest a less pronounced influence on these processes exists in the yeast peroxisomes and mitochondrion compared with humans. However, the dependency of annotation availability on published scientific literature and predictive model development may simply mean that the process in yeast has not been fully experimentally characterized for the information to be readily comparable. It is also feasible that curator judgment and interpretation of the data may play a role in some differences observed in comparison of the yeast and human dataset.
Comparison between biological processes enriched only in human peroxisome proteins (A) and those enriched only in the yeast peroxisomal protein set (B).
Despite some differences in term enrichment, the majority of GO annotations in yeast and human show some similarity in the majority of their biological processes such ‘as cell aging’, ‘intracellular transport’, and ‘long chain fatty acid metabolism’. These similarities support the use of the yeast model by researchers to gain an understanding of peroxisome functions in human.
Discussion
Databases containing concise well-curated records that are annotated consistently using a well-defined and structured vocabulary in a computer-friendly format are invaluable for the quick retrieval of experimental information, facilitating speedy data analysis. It is with this in mind that we selected to focus on curating the full set of human peroxisomal proteins using the GO in order to provide a dataset with easily retrievable information that could be important in studying organelle function.
We have defined a human peroxisomal annotation set using the GO vocabulary. From this effort, 88 of the initial 126 proteins were confirmed peroxisomal by a published direct experimental assay (evidence code ‘Inferred from Direct Assay, IDA’). To date, this is the only publicly available peroxisomal protein list separating experimentally determined and predicted subcellular localization information for this organelle that also provides the supporting citations.
PeroxisomeKB has 101 human peroxisomal gene products, whereas PeroxisomeKB maps 129 concepts of the human peroxisome. The initial 126 proteins that we identified for investigation encompass those found in the two databases plus a few others predicted in the UniProt set. Proteins like TTC1 (UniProtKB Q99614) and VIM (UniProtKB P08670) are listed only in the UniProtKB set as peroxisomal. Our work goes a step further by differentiating those bas062proteins that have experimental evidence for peroxisomal localization compared with those that are only predicted to be peroxisomal.
Limitations of predictive methods of localization
Erroneous prediction due to motif identification
Particular protein motifs widely used as peroxisomal localization determinants can result in false positive annotation statements. In the case of human peroxisome protein import, two distinct sequence features involved in peroxisomal matrix targeting are a C terminal S/A-K/R-LM/ amino acid sequence known as the peroxisomal targeting sequence 1 (PTS1) (27) and an N-terminal sequence (R/K)-(L/V/I)-(XXXXX)-(H/Q)-(L/A/F) designated peroxisomal targeting sequence 2 (PTS2) (28). However sophisticated, sequence considerations cannot eliminate all peroxisomal false positives as target sequence functionality has been shown to be dependent on numerous factors, including protein conformation and targeting sequence accessibility. Other proteins with a fully functional PTS will be targeted to peroxisomes in some tissues or cells and not in others. An example is soluble epoxide hydrolase that localizes to the peroxisomes of hepatocytes and the proximal tubule but in tissues like the adrenal gland, the protein is exclusively cytosolic (29). In contrast, other proteins are able to gain entry into an organelle via chaperone-mediated pores or are localized via piggy back transport (30). This knowledge has steered the curation project to define the human peroxisome proteome as being comprised of proteins with experimental evidence for peroxisomal localization.
Erroneous prediction from sequence-based assumptions
Propagation of peroxisomal localization based on orthology assumptions has additionally been found to have the potential of creating erroneous annotation. The genera Giardia, Trichomonas and Entamoeba do not contain peroxisomes (31), however, some protein sequences from these genera contain sequence motifs that are often used as indicators of peroxisomal localization. UniProt has worked to circumvent such misprediction by the continuous development of rules based on using annotations created with experimental evidence to guide the predictive pipelines output. Details of this and other taxonomy-based rules are available at http://www.ebi.ac.uk/QuickGO/AnnotationPostProcessing.html.
Within the human peroxisome set, we have also encountered well-studied gene products like ACAD11 (UniProtKB Q709F0), ZADH2 (UniProtKB Q8N4Q0), ISOC1 (UniProtKB Q96CN7), TMEM135 (UniProtKB Q86UB9), SOD1 (UniProtKB P00441) and PXT1 (UniProtKB Q8NFP0), which have been predicted by this effort (using the evidence code ‘ISS, Inferred from Sequence Similarity’) to have peroxisomal location from evidence found in rat and mouse orthologs. These annotations were made based on the human proteins having sequence identity >80% (>90% of the length of the proteins) to a protein that has experimental evidence for peroxisomal location. Such proteins with probable peroxisomal location via sequence similarity represent proteins that would benefit from experimental work to define their localization with the predictive method providing some guidance as to their anticipated localization.
Benefits of a peroxisome-focused annotation project
An annotation initiative like the one we have described, provides a distinct enhancement for analysis of protein sets. Annotating with a focus to fully describe all proteins found in an organelle ensures that proteins that may have been neglected in other annotation contexts are treated as priority. An example of the benefits of the annotation effort at an individual protein level is for the insulin degrading enzyme, which is now associated with biologically relevant terms such as ‘determination of adult lifespan’ and ‘bradykinin catabolic process’.
We have also shown the extreme usefulness of predictive annotation sets to provide some biological information where none is available, thus highlighting the importance of using the full complement of GO annotation to understand the functions of an organelle.
The ability to capture in a manual GO annotation contextual data for a protein’s functional roles, such as cell and tissue type or organism developmental stage, will be a great added benefit to future analyses of datasets. As more curation groups start to use this ‘annotation extension’, we will begin to see a more complete picture of the functional interplay between proteins.
Acknowledgements
We thank Pablo Porras-Millán and Sandra Orchard for assistance with protein–protein interaction network advice. Special thanks to the GO editors for the timely provision of terms requested throughout this initiative.
Funding
European Molecular Biology Laboratories; National Institutes of Health (grant no. R01HG02273-02: Gene Ontology Consortium, 2U01HG02712-04: UniProtKB Consortium). Funding for open access charge: European Molecular Biology Laboratory (EMBL)
Conflict of interest. None declared.