-
PDF
- Split View
-
Views
-
Cite
Cite
Jiao Li, Yueping Sun, Robin J. Johnson, Daniela Sciaky, Chih-Hsuan Wei, Robert Leaman, Allan Peter Davis, Carolyn J. Mattingly, Thomas C. Wiegers, Zhiyong Lu, BioCreative V CDR task corpus: a resource for chemical disease relation extraction, Database, Volume 2016, 2016, baw068, https://doi.org/10.1093/database/baw068
Close - Share Icon Share
Abstract
Community-run, formal evaluations and manually annotated text corpora are critically important for advancing biomedical text-mining research. Recently in BioCreative V, a new challenge was organized for the tasks of disease named entity recognition (DNER) and chemical-induced disease (CID) relation extraction. Given the nature of both tasks, a test collection is required to contain both disease/chemical annotations and relation annotations in the same set of articles. Despite previous efforts in biomedical corpus construction, none was found to be sufficient for the task. Thus, we developed our own corpus called BC5CDR during the challenge by inviting a team of Medical Subject Headings (MeSH) indexers for disease/chemical entity annotation and Comparative Toxicogenomics Database (CTD) curators for CID relation annotation. To ensure high annotation quality and productivity, detailed annotation guidelines and automatic annotation tools were provided. The resulting BC5CDR corpus consists of 1500 PubMed articles with 4409 annotated chemicals, 5818 diseases and 3116 chemical-disease interactions. Each entity annotation includes both the mention text spans and normalized concept identifiers, using MeSH as the controlled vocabulary. To ensure accuracy, the entities were first captured independently by two annotators followed by a consensus annotation: The average inter-annotator agreement (IAA) scores were 87.49% and 96.05% for the disease and chemicals, respectively, in the test set according to the Jaccard similarity coefficient. Our corpus was successfully used for the BioCreative V challenge tasks and should serve as a valuable resource for the text-mining research community.
Database URL : http://www.biocreative.org/tasks/biocreative-v/track-3-cdr/
Introduction
Relations between chemicals and diseases (Chemical-Disease Relations or CDRs) play critical roles in drug discovery, biocuration, drug safety, etc. ( 1 ). Because of their critical significance, CDRs are being manually curated by resources such as the Comparative Toxicogenomic Database (CTD; http://ctdbase.org ) ( 2 , 3 ). Due to the high cost of manual curation and rapid growth of the biomedical literature, several attempts have been made to assist curation using text-mining systems ( 4 , 5 ) including the automatic extraction of CDRs ( 6 ). These attempts have met with limited success, however, due in part to the lack of a large-scale training corpus. Through BioCreative V in 2015, one of the major formal evaluations for text-mining research ( 7 ), a new challenge was organized to advance the state-of-the-art in extraction of CDRs ( 8 ). The challenge included two subtasks: disease named entity recognition (DNER) task and chemical-induced disease (CID) relation extraction task.
To support both tasks, a text corpus of PubMed abstracts containing annotations of both chemical/diseases and their interactions is desirable. Despite the existence of many biomedical corpora (see ( 9 ) for a brief review) including a few specifically targeting diseases ( 10–12 ) and chemicals ( 13 ), there were none that fulfilled the following content criteria: (i) inclusion of instances of chemical-disease relation annotations that are asserted from both within and across sentence boundaries; (ii) abstracts containing complete chemical, disease and relation annotations; (iii) chemical/disease annotations grounded in concept identifiers via a controlled vocabulary. Thus, we proposed building a new corpus that satisfies these three requirements.
The proposed corpus is related to some previous efforts in corpus annotation for biomedical information extraction research, such as protein–protein interaction ( 14 ) and drug-drug interaction ( 15 ). It is also significantly different from the previously constructed corpora for mining adverse drug reaction/effects in terms of the annotation scope (CID relations), requirements (see above) and size (1500 articles). As shown in Table 1 , the EU-ADR corpus contains a total of 300 PubMed articles with 739 drugs, 812 diseases and 300 drug-disease associated relations at sentence level ( 16 ). The ADE corpus consists of 2972 PubMed articles with sentence-level statements of 5776 adverse effects related to 5063 drugs ( 17 ). The corpus developed by ( 18 ) served for disease and adverse effect named entity recognition tasks rather than relation extraction.
Comparison with the previous chemical disease relation corpora
| Corpus . | Annotation scope . | Size . | Entity annotation—Mention . | Entity annotation—Concept . | Relation annotation . |
|---|---|---|---|---|---|
| BC5CDR | Abstract | 1500 | Yes | Yes | Yes |
| EU-ADR (16) | Sentence | 300 | Yes | Yes | Yes |
| ADE (17) | Sentence | 2972 | Yes | No | Yes |
| Corpus (18) | Abstract | 400 | Yes | Yes | No |
| Corpus . | Annotation scope . | Size . | Entity annotation—Mention . | Entity annotation—Concept . | Relation annotation . |
|---|---|---|---|---|---|
| BC5CDR | Abstract | 1500 | Yes | Yes | Yes |
| EU-ADR (16) | Sentence | 300 | Yes | Yes | Yes |
| ADE (17) | Sentence | 2972 | Yes | No | Yes |
| Corpus (18) | Abstract | 400 | Yes | Yes | No |
Comparison with the previous chemical disease relation corpora
| Corpus . | Annotation scope . | Size . | Entity annotation—Mention . | Entity annotation—Concept . | Relation annotation . |
|---|---|---|---|---|---|
| BC5CDR | Abstract | 1500 | Yes | Yes | Yes |
| EU-ADR (16) | Sentence | 300 | Yes | Yes | Yes |
| ADE (17) | Sentence | 2972 | Yes | No | Yes |
| Corpus (18) | Abstract | 400 | Yes | Yes | No |
| Corpus . | Annotation scope . | Size . | Entity annotation—Mention . | Entity annotation—Concept . | Relation annotation . |
|---|---|---|---|---|---|
| BC5CDR | Abstract | 1500 | Yes | Yes | Yes |
| EU-ADR (16) | Sentence | 300 | Yes | Yes | Yes |
| ADE (17) | Sentence | 2972 | Yes | No | Yes |
| Corpus (18) | Abstract | 400 | Yes | Yes | No |
Methods and materials
Article selection
We selected a total of 1500 articles for the CDR task, split into three subsets: 500 each for the training, development and test sets. The training, development and most (400) of the test set were randomly selected from the CTD-Pfizer corpus, which was generated via a previous collaboration between CTD and Pfizer, and comprises over 150 000 chemical-disease relations from 88 000 articles ( 19 ).
To ensure we have some unseen data for the task participants, the remaining 100 articles of the test set were annotated during the challenge (i.e. not selected from the previous CTD-Pfizer corpus) and their curation was not made public until the BioCreative V challenge was complete. We used the following method to select the 100 articles to ensure they would have a similar distribution of words as the training and development sets. For each of the 1000 articles in the training and development sets, we retrieved the list of related articles using PubMed E-utilities. We removed from consideration any articles that did not meet our selection criteria. Specifically, the target article must be in English, contain an abstract, and be published in 2014 or later. For each new article, we computed an overall score by summing the similarity scores ( 20 , 21 ) between the target article and each article in the training and development sets. We also determined an overall similarity score for each article in the training and development sets with a similarity score calculated using all other articles in the training and development sets. We then selected the final set by sampling with replacement from the similarity distribution of the training and development sets: we randomly selected an article from the training or development sets, obtained its similarity score, and then selected the new article with the closest similarity score. The resulting articles are approximately as well related to the articles in the training and development sets, in terms of similarity scoring, as the articles in the training and development sets are related to one another.
Annotation tasks
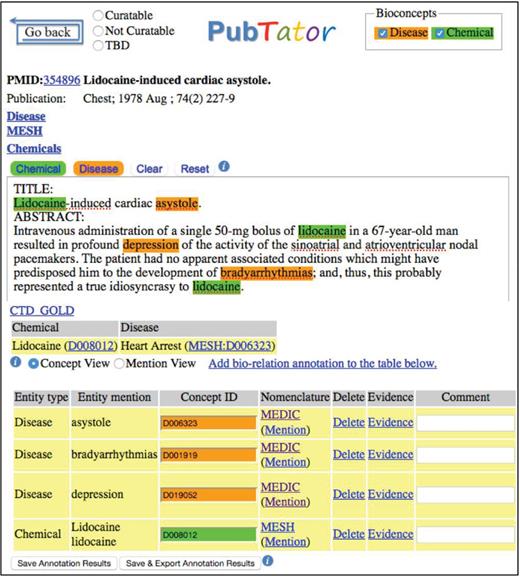
We performed manual annotation of all chemicals and diseases mentioned in the 1500 articles. For each entity occurrence, we not only annotated its text span but also assigned a relevant concept identifier from MeSH ( 22 ). As shown in Figure 1 , three diseases mentioned in the abstract were highlighted by our automated tool for potential consideration by the MeSH annotators, along with three occurrences of the same chemical (Lidocaine).
As indicated above, we largely leveraged the previous annotation of chemical-disease relationships from the CTD-Pfizer dataset for 1400 of the 1500 articles with few changes: (i) we removed relations that required entities not found in abstracts; (ii) we removed relations that were not disease specific (e.g. ‘Drug-Related Side Effects and Adverse Reactions’ (D064420)); and (iii) we updated a few CTD relations due to the MeSH vocabulary changes (the CTD-Pfizer project was conducted in years 2011/12, and the MeSH vocabulary has changed since then).
We performed new manual annotation of chemical–disease relations for the remaining 100 articles in the test set. For the BioCreative V challenge task, the CID relations refer to two types of relationships between a chemical and a disease in CTD:
Putative mechanistic relation between a chemical and a disease indicates that the chemical may play a role in the etiology of the disease (e.g. exposure to chemical X causes lung cancer). Figure 1 shows an example of such a CTD curated relationship between Lidocaine and Heart Arrest (disease term for the synonym ‘asystole’ used by the authors in the abstract).
Biomarker relation between a chemical and a disease indicates that the chemical correlates with the disease (e.g. increased abundance in the brain of chemical X correlates with Alzheimer disease).
CTD curators used their standard curation process for CDR curation ( 23 ). Curation was limited to the title and abstract except in cases where reference to the full text was required for clarification; abstracts that required full-text curation were removed from the corpus. In addition to CDR curation, all observed interactions and relationships applicable to CTD were curated for each abstract. CTD triaged and/or curated 143 articles in conjunction with BioCreative V; the final 100 selected for inclusion in the Test Dataset represented abstract-only curation for CDRs.
Annotators
For entity annotation, we recruited four MeSH indexers, all of whom had a medical training background and curation experience. Each article was annotated independently by two annotators (i.e. double-annotation). Differences were resolved by a third and senior annotator (YS). Three CTD annotators curated the relationships between chemicals and diseases.
Annotation guidelines
The task organizers followed the usual practice of biomedical corpus annotation for entity annotation: the MeSH annotators were asked to follow an initial set of guidelines when annotating the first 100 sample articles. Annotation discrepancies and questions were discussed and settled by the senior annotator; the annotation guidelines were revised accordingly. Detailed guidelines are available on the task website. For CID relation annotation, the standard CTD curation protocol was followed ( 23 ).
Annotation tools
Manual annotation of disease and chemical entities was performed using PubTator ( 4 , 5 ) (see Figure 1 ). To accelerate manual annotation ( 24 ), text-mined disease and chemical results were pre-computed using DNorm ( 25 ) and tmChem ( 26 ) and displayed to the annotators. When necessary, the annotators added new annotations, and deleted or edited the automatic annotations based on their judgment. The annotators were permitted to use public resources such as UMLS or Wikipedia to facilitate the annotation process. CTD’s in-house Curation Tool ( 23 ) was used for all relation curation.
Annotation data formats
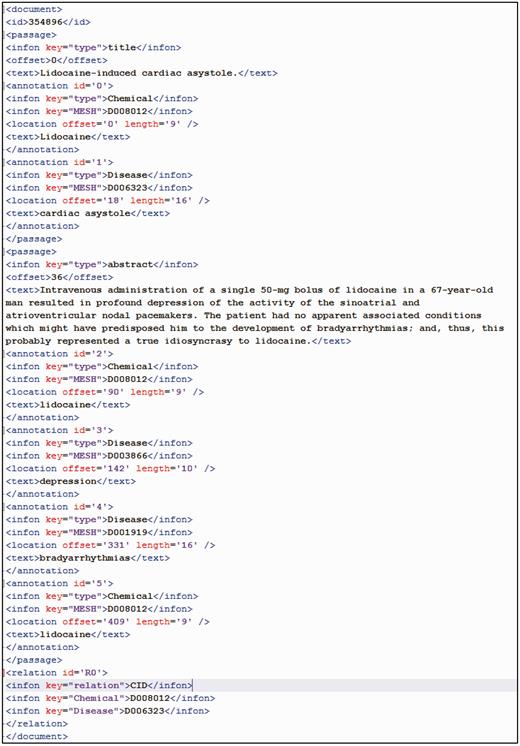
All annotated data were made available to participants in both PubTator and BioC formats. The PubTator format consists of a straightforward tab-delimited text file. Figure 2 shows the tab-delimited file for the article (PMID: 354896) in training set. The BioC ( 27 ) format is an XML standard recently proposed for biomedical text-mining and data output. For the same article, its BioC format is shown in Figure 3 .
Inter-annotator agreement (IAA) analysis
IAA for CTD relation curation was previously described in ( 29 ); no further experiments were conducted in this study.
Results and discussion
Corpus overview
The corpus consists of three separate sets of articles with diseases, chemicals and their relations annotated. The training (500 articles) and development (500 articles) sets were released to task participants in advance to support text-mining method development. The test set (500 articles) was used for final system performance evaluation. As shown in Table 2 , the three data sets have similar distributions of chemical mentions, disease mentions and CID relations, which makes the corpus more useful for training models. The table also shows that while there are more chemical mentions than disease mentions in the corpus, there are more disease entities (IDs) than chemical entities (IDs).
The overall corpus statistics
| Task dataset . | Articles . | Disease . | Chemical . | CID relation . | ||
|---|---|---|---|---|---|---|
| Mention . | ID . | Mention . | ID . | |||
| Training | 500 | 4182 | 1965 | 5203 | 1467 | 1038 |
| Development | 500 | 4244 | 1865 | 5347 | 1507 | 1012 |
| Test | 500 | 4424 | 1988 | 5385 | 1435 | 1066 |
| Task dataset . | Articles . | Disease . | Chemical . | CID relation . | ||
|---|---|---|---|---|---|---|
| Mention . | ID . | Mention . | ID . | |||
| Training | 500 | 4182 | 1965 | 5203 | 1467 | 1038 |
| Development | 500 | 4244 | 1865 | 5347 | 1507 | 1012 |
| Test | 500 | 4424 | 1988 | 5385 | 1435 | 1066 |
The overall corpus statistics
| Task dataset . | Articles . | Disease . | Chemical . | CID relation . | ||
|---|---|---|---|---|---|---|
| Mention . | ID . | Mention . | ID . | |||
| Training | 500 | 4182 | 1965 | 5203 | 1467 | 1038 |
| Development | 500 | 4244 | 1865 | 5347 | 1507 | 1012 |
| Test | 500 | 4424 | 1988 | 5385 | 1435 | 1066 |
| Task dataset . | Articles . | Disease . | Chemical . | CID relation . | ||
|---|---|---|---|---|---|---|
| Mention . | ID . | Mention . | ID . | |||
| Training | 500 | 4182 | 1965 | 5203 | 1467 | 1038 |
| Development | 500 | 4244 | 1865 | 5347 | 1507 | 1012 |
| Test | 500 | 4424 | 1988 | 5385 | 1435 | 1066 |
Inter-annotator agreement for mention annotation
Table 3 shows the inter-annotator agreement (IAA) scores of three separate subsets for both disease and chemical annotations. The IAA scores over the entire corpus are 87.49% (diseases) and 96.05% (chemicals), which suggests higher agreement on chemical than disease mentions. Additionally, the IAA scores are slightly higher on the test set than the training and test sets.
Inter-annotator agreement (IAA) scores of the three sets
| Task dataset . | Disease . | Chemical . |
|---|---|---|
| Training | 0.8600 | 0.9523 |
| Development | 0.8742 | 0.9577 |
| Test | 0.8875 | 0.9630 |
| All | 0.8749 | 0.9605 |
| Task dataset . | Disease . | Chemical . |
|---|---|---|
| Training | 0.8600 | 0.9523 |
| Development | 0.8742 | 0.9577 |
| Test | 0.8875 | 0.9630 |
| All | 0.8749 | 0.9605 |
Inter-annotator agreement (IAA) scores of the three sets
| Task dataset . | Disease . | Chemical . |
|---|---|---|
| Training | 0.8600 | 0.9523 |
| Development | 0.8742 | 0.9577 |
| Test | 0.8875 | 0.9630 |
| All | 0.8749 | 0.9605 |
| Task dataset . | Disease . | Chemical . |
|---|---|---|
| Training | 0.8600 | 0.9523 |
| Development | 0.8742 | 0.9577 |
| Test | 0.8875 | 0.9630 |
| All | 0.8749 | 0.9605 |
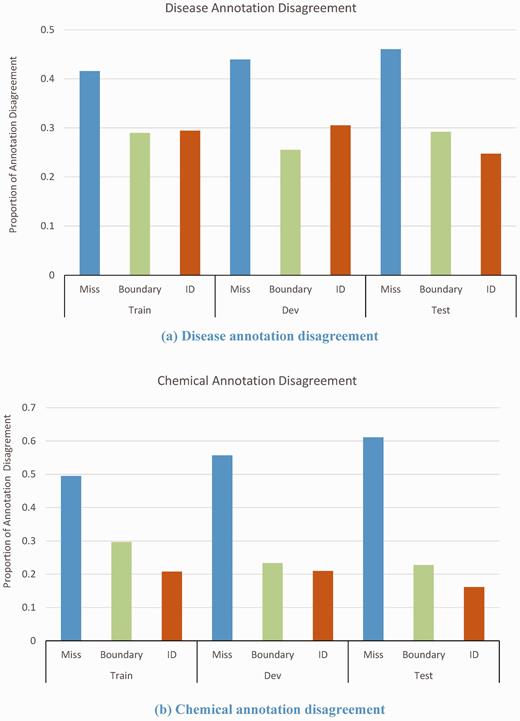
Analyzing the disagreements, we found that many were caused by missed annotation, boundary disagreement and inconsistent identifier assignment. Figure 4 shows the proportion of disease and chemical annotation disagreements in the training, development and test sets, respectively. The most of disagreements (∼50%) were due to missed annotation, where one annotator failed to identify the disease/chemical mentions recognized by the other. 28% of disease annotation disagreements were related to the boundary issue. For example, in the article (PMID 20466178) entitled ‘Rosaceiform dermatitis associated with topical tacrolimus treatment’, it was difficult to judge whether annotate ‘rosaceiform dermatitis’ as ‘rosacea’ (MeSH ID: D012393) or simply annotate ‘dermatitis’ (MeSH ID: D003872).
There were also many cases of disagreement over the concept identifier of diseases, especially for the mentions where the text did not exactly match any MeSH term. In some cases, it was hard to judge whether to assign an unknown concept identifier of ‘-1’ or an ancestor concept identifier. For example, in the article with PMID of 12093990, one annotator selected ‘infection with hemorrhagic fever viruses’ as ‘-1’, while the other selected ‘D006482’ (Hemorrhagic Fevers, Viral). In this case the adjudicating annotator chose the latter term.
Disease mention distribution
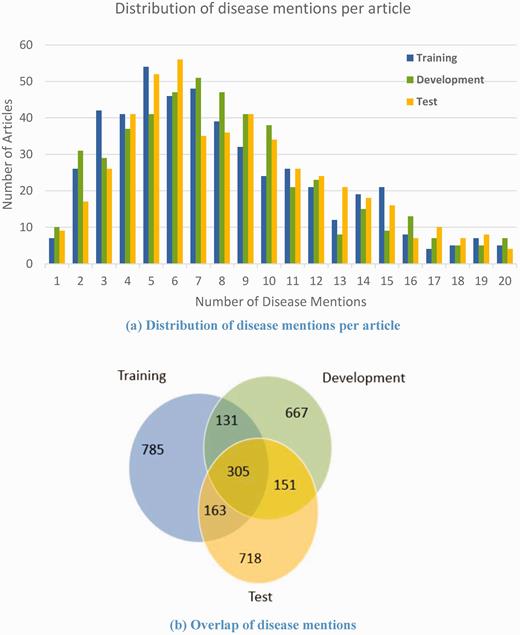
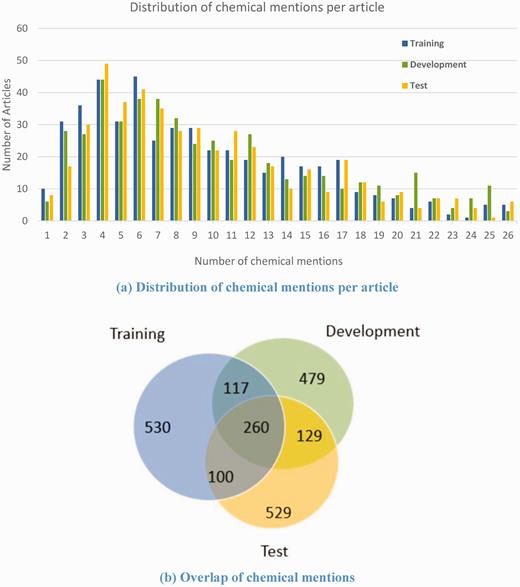
On average, the corpus contains 8.57 non-distinct disease mentions per PubMed abstract. Figure 5(a) shows the breakdown of the number of disease mentions per article in the training, development and test sets, respectively. The three data sets have similar disease mention distribution. In addition, we compared the overlap of unique disease mentions in the three data sets as shown in Figure 5(b) . It can be seen that 718 disease mentions out of 1337 in the test set never appear in the training set or the development set. The similar distribution and differential disease mentions in the three sets make the corpus more useful for training models.
Chemical mention distribution
Compared with diseases, the corpus contains more non-distinct chemical mentions (10.62) per PubMed abstract on average. The chemical mention distribution in the three sets ( Figure 6(a) ) and the overlap of unique chemical mentions in the three sets ( Figure 6(b) ) demonstrate that, like the disease mentions, the corpus has excellent characteristics to support model training for chemical entity recognition. Here, we used MeSH identifiers to normalize the chemical mentions because CID relations were previously annotated already in MeSH which is designed for literature indexing and has been used in similar annotation projects ( 26 , 30 ). The CTD chemical vocabulary ( http://ctdbase.org/downloads/#allchems ) can facilitate mapping the MeSH identifiers to other chemical resource accessions for further chemical-related studies.
CID relation distribution
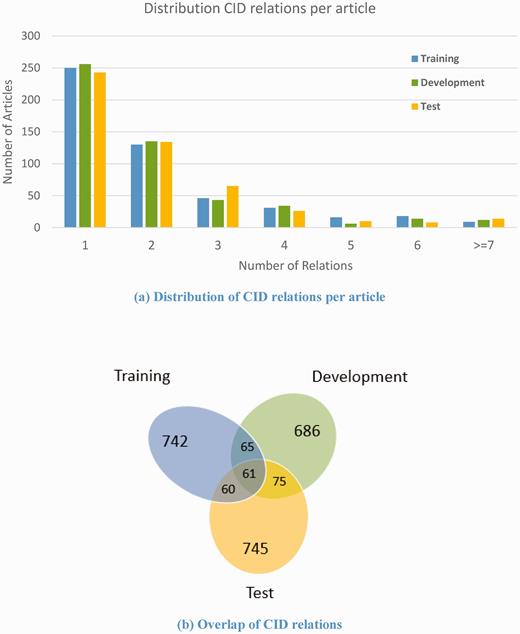
For CID relationships, the average number of relations per PubMed abstract is 2.08. About 50% of articles in the corpus have only one CID relation per article, and 86.8% of articles have no more than three relations ( Figure 7(a) ). Unlike the disease and chemical mentions, the overlap of unique CID relations in the three sets is as low as 61 relations; 79.17% (745 out of 941) of the relations in the test set have never appeared in the training or development set ( Figure 7(b) ). This makes relation extraction task more challenging.
Distribution of chemical-induced disease relations in the corpus.
Corpus usage in BioCreative V
The corpus successfully supported the BioCreative V Chemical Disease Relation (CDR) task ( 8 , 31 ). A total of 34 teams worldwide participated in the task: 16 teams participated in the in the DNER task, and 18 teams participated in the CID task. As reported in the BioCreative V, the best system performance F-score was 86.46% for the DNER task ( 32 ) and 57.03% for the CID task ( 33 ). This corpus provides a benchmark set to facilitate further improvement for biomedical text-mining method development, especially as it relates to semantic relationship extraction.
Conclusions
We developed a corpus for both named entity recognition and chemical-disease relations in the literature. A total of 1500 articles have been annotated with automated assistance from PubTator. Jaccard agreement results and corpus statistics verified the reliability of the corpus. Furthermore, our annotated data includes the CDR relations that are asserted across sentence boundaries (i.e. not in the same sentences). We believe this data set will be invaluable for advancing text-mining techniques for relation extraction tasks.
Acknowledgment
We would like to thank Yifan Peng for his help on the manuscript.
Funding
The research was supported by the National Population and Health Scientific Data Sharing Program of China, the China Knowledge Centre for Engineering Sciences and Technology (Medical Centre), the Fundamental Research Funds for the Central Universities (Grant No. 13R0101), the National Institute of Environmental Health Sciences (ES014065 and ES019604). Funding for open access charge: The National Institutes of Health Intramural Research Program.
Conflict of interest . None declared.
References
PubMed Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US);
Author notes
Citation details: Li,J., Sun,Y., Johnson,R.J. et al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database (2016) Vol. 2016: article ID baw068; doi:10.1093/database/baw068