-
PDF
- Split View
-
Views
-
Cite
Cite
Lope A. Flórez, Sebastian F. Roppel, Arne G. Schmeisky, Christoph R. Lammers, Jörg Stülke, A community-curated consensual annotation that is continuously updated: the Bacillus subtilis centred wiki SubtiWiki, Database, Volume 2009, 2009, bap012, https://doi.org/10.1093/database/bap012
Close - Share Icon Share
Abstract
Bacillus subtilis is the model organism for Gram-positive bacteria, with a large amount of publications on all aspects of its biology. To facilitate genome annotation and the collection of comprehensive information on B. subtilis, we created SubtiWiki as a community-oriented annotation tool for information retrieval and continuous maintenance. The wiki is focused on the needs and requirements of scientists doing experimental work. This has implications for the design of the interface and for the layout of the individual pages. The pages can be accessed primarily by the gene designations. All pages have a similar flexible structure and provide links to related gene pages in SubtiWiki or to information in the World Wide Web. Each page gives comprehensive information on the gene, the encoded protein or RNA as well as information related to the current investigation of the gene/protein. The wiki has been seeded with information from key publications and from the most relevant general and B. subtilis-specific databases. We think that SubtiWiki might serve as an example for other scientific wikis that are devoted to the genes and proteins of one organism.
Database URL: The wiki can be accessed at http://subtiwiki.uni-goettingen.de/
Introduction
With the completion of more and more genome sequences, their accurate annotation has become an important matter. Usually, the initial annotation is done automatically, and is subsequently improved by manual curation. All major model organisms that are subject to extensive investigation have been sequenced and annotated in the early phase of the genomic age. However, once a genome sequence and the corresponding annotation have been published, there is decreasing support for and interest in keeping the annotation information up-to-date. Since the work in the ‘traditional’ molecular and cell biology labs goes on, new information is continuously being generated but not included in the annotation. A good example for this problem is the small RNA SR1 of the bacterium Bacillus subtilis that was originally described in 2005, but that is not annotated even in the most recent publication of the B. subtilis genome (1,2). Since experimental work focuses on a few model organisms, this problem is of specific urgency for these organisms. At the same time, the lack of complete up-to-date annotation information may prevent the lab researchers from getting important new insights because the relevant information is not easily accessible from primary literature. The problem of outdated annotation is even aggravated by the fact, that annotation for one organism is usually controlled by one institution that might change its focus and thus be unable to guarantee updated annotation in the long term.
A way to overcome these problems might be to establish an annotation based on the wiki concept. This concept offers several advantages: first, each interested scientist can easily contribute any information to the existing annotation and make it thus more useful. The result is that novel information can be added immediately upon its generation and its inclusion does not depend on the availability of a usually unknown curator. Thus, the task of annotation is distributed among a complete scientific community. Second, a wiki makes it very easy to retrieve complete sets of object-oriented information, which are represented as a wiki page. Moreover, the information provided can be enriched by internal and external links to pages of the wiki and the internet, respectively. These links establish different classes of connections that make the interrelatedness of all processes of life visible and tractable. Third, a wiki is a liberal way to manage shared information. Alternative opinions can be exchanged and presented as such without somebody who has the power to decide what the truth is. Instead, each user can make its own judgement and assess the validity of opposing statements not only based on the evidence provided but also on the own additional knowledge.
Compared to classical relational databases, a wiki has some similarities but there are also fundamental differences. Both provide the user with the requested information. However, whereas the structure of a relational database is very rigid, a wiki can be very flexible. In principle, each page could have an individual structure that is adjusted to the information to be presented on the page. This may cause problems if one wants to extract an identical set of information from each page of the wiki. However, since the wiki is object-centred, it can be especially successful for the retrieval of information on individual objects such as genes or proteins. In contrast, relational databases outcompete wikis for the retrieval of cross-sectional information. The more flexible structure of wiki pages allows the presentation of information to a level of detail that is unprecedented in relational databases. The simple structure of the wiki pages and the inherent user-friendliness make the wiki an easy-to-access and easy-to-contribute marketplace of information.
These advantages of the wiki concept resulted in a large number of different kinds of scientific wikis that have been established in the past few years resulting in the new ‘discipline’ of ‘wikiomics’ (3). Wikis have been set up for different biological purposes such as ArrayWiki for the annotation of microarray experiments (4), Proteopedia for protein structures (5) and EcoliWiki for the model bacterium Escherichia coli (6). In addition to these more specialized wikis, there are general wikis devoted to all genes and proteins as well as to metabolic pathways (7–9). The wiki concept has been suggested to be of specific value for genome re-annotation due to the challenges mentioned above (10).
We are interested in the Gram-positive model bacterium B. subtilis (11,12). These bacteria are of great practical importance because they are used in biotechnology for the production of vitamins and enzymes (13). Moreover, B. subtilis undergoes a simple differentiation program and is the model to understand many important pathogens such as B. anthracis, Staphylococcus aureus and Listeria monocytogenes. Therefore, B. subtilis has attracted substantial research interest during the past decades that has made this bacterium the best-studied in addition to E. coli (14). The genome sequence of B. subtilis has been published in 1997 (15) and the publicly available annotation has not been updated from 2001 to 2009 (2). In an attempt to facilitate continuous genome annotation, we have set up a wiki devoted to the genes and proteins of B. subtilis. In this wiki, designated SubtiWiki, information is centred on the genes and the corresponding proteins (or RNAs) of B. subtilis. The wiki provides information from mutant phenotypes, gene expression and regulation, to the functions, modifications, interactions and localizations of proteins. Moreover, SubtiWiki provides links to databases specialized in gene expression, genome organization, protein structures and enzyme activities. Finally, the wiki provides information on biological materials, specialists as well as links to relevant publications.
Description of the wiki
The central objects of SubtiWiki are the genes, proteins and functional RNAs of B. subtilis. Thus, most pages of the wiki are devoted to a specific gene and its corresponding product(s). The central position of the genes is indicated by a search box on the start page of SubtiWiki, which can be used to enter the name of the gene of interest, to get access to detailed information on this gene. Moreover, information can be retrieved by text search through all pages of the wiki. Both the gene pages and the main page provide links to other categories of pages such as pages for the labs that work with B. subtilis, or pages for important plasmids and methods (see below).
Gene names as identifiers
There are two principal options to get access to gene-specific pages. One would be to use genetic gene designations, whereas the alternative is the use of gene identifiers derived from genomic projects. The latter option is preferable for organisms in which only a small part of the genes had been studied before, and where annotations are therefore not yet stably established in the scientific literature. In contrast, B. subtilis has been the object of substantial investigation since the middle of the last century, and this interest is going on, and has become even more intensive, with the availability of the genome sequence (14). The use of classical gene designations has a long-standing tradition in the work with B. subtilis, and it is safe to predict that each Bacillus researcher knows the designations of at least 100 genes together with the corresponding products and functions. With such a strong tradition, and with the needs of the scientific Bacillus community in mind, we decided to build the pages on the gene names. This brings of course the problem of instability of certain designations and of synonyms. Based on a collection of these synonyms that is available in our group, we ensured that all common designations of a gene guide the user to the same gene page via redirects. This is the case for 559 genes that represent about 12% of all B. subtilis genes. With the ongoing research and the identification of new gene functions and the introduction of novel mnemonic designations, more redirects are likely to be required.
However, gene identifiers are the most stable way to refer to specific genes and proteins since they do not change with the accumulation of novel information. The standard gene identifiers for B. subtilis genes are the identifiers provided by the annotation team at the Institut Pasteur (2,15). These identifiers are mapped to the gene designations used in SubtiWiki and the Uniprot identifiers in an Excel table. This table is available on the front page of SubtiWiki.
Semi-structured pages balance intuitive orientation with the ease of contributing
The structure of the pages of scientific wikis may be very different. On one end of the scale are WikiGenes and Proteopedia, which use text descriptions. This may make it difficult to find the requested information on a page since every page may present the content differently. On the other end, the information in EcoliWiki is entered in tables with a rigid structure. Such a structure is very helpful to achieve consistency in the presented information and in the design of the pages. At the same time it poses two problems: first, for some data there may be no obviously appropriate table. Second, and perhaps even more serious, such a structure might discourage the casual user from contributing information. However, the wiki is aimed specifically at such users that generate new experimental information and add it to the wiki without training in the curation of a wiki.
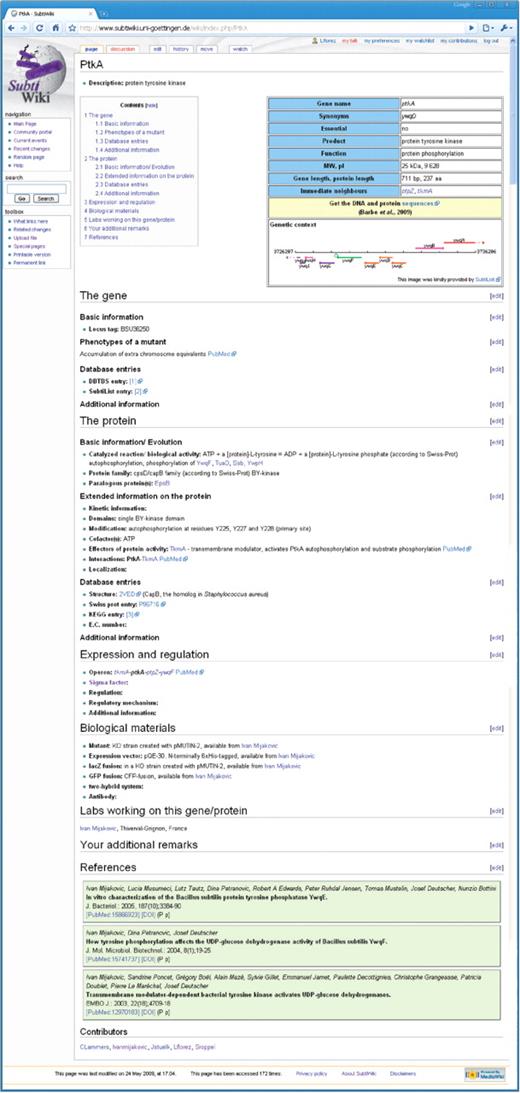
For SubtiWiki, we decided to strike a balance between the two extreme strategies outlined above. The most critical information that is required very often is presented in a table at the top of each page (Figure 1). All additional information is provided as text under preset headlines. These headlines are listed in a Table of Contents on the top of each page next to the table with the key information. This general outline is derived from a template page that was used to generate all the individual gene pages. While the structure of the table is quite rigid, all other headlines can be easily adapted, irrelevant headlines can be deleted and new headlines be added. With these possibilities, each page can be adapted to the specific requirements of the gene and its products although the general layout of all pages remains still very similar. This makes it very easy for the user to go directly to the set of information he/she is interested in. Moreover, this way of arranging the pages is very advantageous for the addition of information: the common general headlines facilitate not only the automatic entry of information using scripts, but it is also very easy for the user to edit the information since an ‘edit’ button is present next to each headline, and new contents can be added intuitively. We are confident that our page layout will lower the barrier for the casual user.
The layout of gene pages in SubtiWiki. The pages adhere to the design used in Wikipedia. At the top, the user finds a clickable table of contents of the detailed information. Next to it there is another table with the most important information on a gene/protein and a scheme of the genomic context (see Figure 2 for details). These tables are then followed by detailed information on the gene, the protein or RNA and gene expression/regulation (see Figure 3 for details). The next sections cover biological materials related to the gene/protein, the labs working on the gene or protein, and provide space for additional remarks, which do not seem to fit elsewhere on the page. Finally, references and information on the contributors are listed (see Figure 4 for details).
Features of the pages for the individual genes
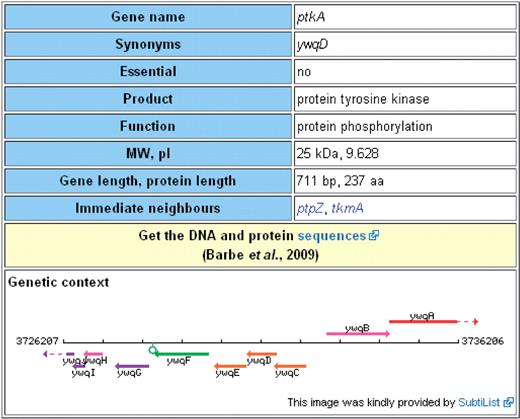
As mentioned above, at the top of each gene page there is the Table of Contents for the detailed information provided in the bottom part of the page as well as a table with the most important information on a gene (Figure 2). This table contains information on gene designations and synonyms, the gene product and its function, whether the gene is essential or not, quantitative information important for the experimental work (gene and protein length, molecular weight and isoelectric point of the protein) and the gene context (the neighbouring genes and a figure showing the context). Moreover, this table provides a link to the DNA and amino acid sequences entry in the EMBL Nucleotide Sequence Database (16). In contrast to the detailed information for the genes, which will remain work in progress as long as the research on B. subtilis continues, the table with the key information has been completed for all genes (for the source of information, see below).
Key information on any gene/protein. Each page contains a table with information that is most often accessed by experimental biologists. The table provides information on synonyms, states whether a gene is essential or not and gives details on the gene product and its function(s). Moreover, the table provides ‘technical data’ such as lengths of the gene and the corresponding protein as well as the molecular weight and the isoelectric point of the protein. Next, the table contains information on the genomic context (neighbouring genes and a figure with of the 10 kb region). Finally, there is a link that gives access to the DNA and protein sequences.
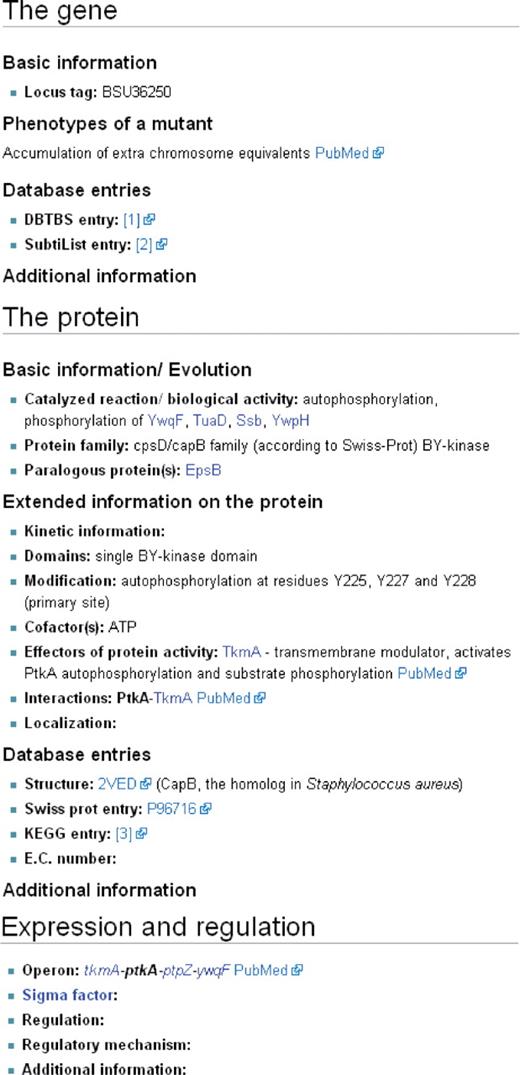
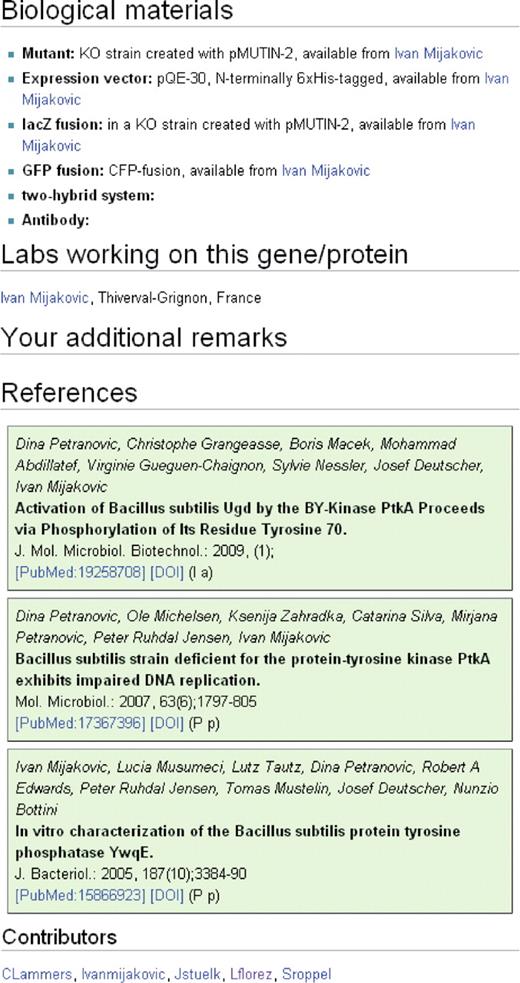
The detailed information is provided in seven categories that belong either to the molecular description of the gene and its product or that are related to the ongoing research on the gene. For the molecular description (Figure 3), there is first a section with information on the gene such as the locus tag, the phenotype(s) of a mutant and links to gene-centred databases. The second section deals with the properties of the encoded protein. The first sub-section covers the biological activity of the protein and evolutionary aspects such as the protein family and paralogous proteins encoded in the genome of B. subtilis. The second sub-section provides detailed information on the protein such as kinetic information, the domain structure, modifications, cofactors and effectors of the biological activity, interactions and the localization of the protein. Finally, the third part of this section contains links to protein-centred databases that cover protein structures and protein activities. The third section providing molecular information is devoted to gene expression and regulation. Here, the operon structures and sigma factors are listed. Moreover, this section provides information on gene regulation and the corresponding regulators and their regulatory mechanisms. To facilitate the research on B. subtilis, the second part of the pages provides information on biological materials and their availability (mutants, expression vectors, GFP fusions, antibodies, etc.), on the labs that work on a gene/protein and the key references on the gene or protein (Figure 4). For all information that does not easily fit into the provided frame, there is a section for additional information. Moreover, such a sub-section is also present in each of the three parts that cover the molecular biology. There are some genes that encode RNAs rather than proteins. For such genes, the page content was adapted accordingly, and the section on the protein was replaced by a section on the RNA.
Biological information on the gene/protein. The pages provide a frame for entering detailed information on any gene or protein. The first section is devoted to the gene and provides information such as the locus tag, phenotypes of mutants and gene-related database entries. The second section covers information on the protein such as the biological activity, the membership in a protein family, and the presence of paralogous proteins in B. subtilis. Moreover, features such as the domain structure, modifications, cofactors and effectors of the biological activity, interaction partners and the protein localization are available. Again, this section ends with protein centred-databases. The third section describes gene expression and regulation. Here, the user finds the operon structure, information on the sigma factor(s) and regulatory mechanisms.
Information on the research on the gene/protein. The first section of this part gives information on biological materials such as mutants, reporter fusions, expression systems or antibodies. The second section shows the labs that study the gene/protein, and there is a section for additional remarks that do not seem to be appropriate at any other position of the wiki. The last section covers the references. At the very bottom of each page, the contributors to this page are shown. These entries are generated automatically.
An important general feature of all gene-specific pages is the availability of external and internal links. External links direct the user to databases or to the publications that describe the data that are presented in the wiki. Internal links relate each gene/protein with all other genes or proteins with which it interacts in one or the other way (physical interaction, regulation or co-localization on the genome). Moreover, the part of the page devoted to the research on a gene provides internal links to pages on resources like plasmids and experimental approaches as well as to pages with information on the labs that study a gene or protein. These features are intended to facilitate the collaboration among the Bacillus labs.
Implementation of SubtiWiki
The content management system
MediaWiki was chosen as the software platform for the wiki (www.mediawiki.org). This interface is identical to that used by Wikipedia, and thus most users can be expected to be immediately familiar with the way of interacting with the system. In addition, MediaWiki allows the use of extensions. Due to the popularity of MediaWiki, many extensions are already available for immediate use. For SubtiWiki, we use three extensions: first, ContributionCredits (http://www.mediawiki.org/wiki/Extension:ContributionCredits) allows giving any contributor a credit for his work by placing his username at the bottom of each page. This is very important for two reasons: on the one hand, the user is acknowledged for each contribution, but on the other hand, this protects the wiki from potential anonymous spam. Second, reCAPTCHA (http://www.mediawiki.org/wiki/Extension:ReCAPTCHA) is used to prevent anonymous automated registration by malicious scripts. This is achieved by requesting the entry of two words upon registration. These words are easily recognized by any person, but they cannot be processed by computer programs (17). Third, the extension Pubmed (http://www.mediawiki.org/wiki/Extension:Pubmed) serves to fetch literature citations from PubMed entries.
User access and restrictions
All pages of SubtiWiki are freely accessible without prior registration. However, the contribution of information is only possible for registered users who are logged in. Our policy with respect to registrations is based on two conflicting aims: on the one hand, we wish to invite users to contribute to the wiki rather than to discourage them by complicated procedures. On the other hand, the reliability of the information provided by SubtiWiki is of crucial importance. A mandatory but liberal registration policy seemed to be the best way to balance these two aims. Thus, registration is simple and does not require the approval of another user. As mentioned above, automated spam registrations are prevented by the reCAPTCHA extension. Once a registered user has logged into the system, any input is possible. With our system of giving credits to all contributors, there is another level of security since nobody is able to modify any pages without being revealed. Moreover, we do not expect vandalism to be a major problem for specialized scientific wikis.
Sources of information
The information provided in SubtiWiki is derived from three principal types of sources. First, we used information from general databases (Table 1). Second, and most importantly, we derived information from databases that are specifically devoted to B. subtilis, and finally, the scientific literature was an important source of information (Table 2). Each page contains information derived from each of these sources because the different databases serve specialized purposes. We did not only extract information from these sources, but we did also provide links to them whenever possible and appropriate. This allows the user to profit from the specific strengths of the individual information sources.
Databases used to seed SubtiWiki
| Database (reference) . | Extracted information, URL . |
|---|---|
| General databases | |
| EMBL-bank (16) | Link to the nucleotide and protein sequences. EC Numbers of enzymes. http://www.ebi.ac.uk/embl/ |
| KEGG (20) | Link to the corresponding gene entry. http://www.genome.jp/kegg/ |
| MPIDB (21) | Protein–protein interactions. http://www.jcvi.org/mpidb |
| PDB (22) | Link to the molecular structures. http://www.pdb.org/ |
| PubMed | Literature citations. http://www.ncbi.nlm.nih.gov/pubmed |
| Swiss-Prot/UniProt (23) | Link to the protein entry. Protein family, localization and catalysed reaction. http://www.uniprot.org/ |
| Databases specific for B. Subtilis | |
| DBTBS (24) | Link to the corresponding operon entry. Operon structure and regulation. http://dbtbs.hgc.jp/ |
| SubtiList (18)/GenoList (25) | Link to the corresponding gene entry. Name, length, product, function and genomic context of the genes. Molecular weight, isoelectric point and length of the protein. http://genolist.pasteur.fr/ |
| Database (reference) . | Extracted information, URL . |
|---|---|
| General databases | |
| EMBL-bank (16) | Link to the nucleotide and protein sequences. EC Numbers of enzymes. http://www.ebi.ac.uk/embl/ |
| KEGG (20) | Link to the corresponding gene entry. http://www.genome.jp/kegg/ |
| MPIDB (21) | Protein–protein interactions. http://www.jcvi.org/mpidb |
| PDB (22) | Link to the molecular structures. http://www.pdb.org/ |
| PubMed | Literature citations. http://www.ncbi.nlm.nih.gov/pubmed |
| Swiss-Prot/UniProt (23) | Link to the protein entry. Protein family, localization and catalysed reaction. http://www.uniprot.org/ |
| Databases specific for B. Subtilis | |
| DBTBS (24) | Link to the corresponding operon entry. Operon structure and regulation. http://dbtbs.hgc.jp/ |
| SubtiList (18)/GenoList (25) | Link to the corresponding gene entry. Name, length, product, function and genomic context of the genes. Molecular weight, isoelectric point and length of the protein. http://genolist.pasteur.fr/ |
Databases used to seed SubtiWiki
| Database (reference) . | Extracted information, URL . |
|---|---|
| General databases | |
| EMBL-bank (16) | Link to the nucleotide and protein sequences. EC Numbers of enzymes. http://www.ebi.ac.uk/embl/ |
| KEGG (20) | Link to the corresponding gene entry. http://www.genome.jp/kegg/ |
| MPIDB (21) | Protein–protein interactions. http://www.jcvi.org/mpidb |
| PDB (22) | Link to the molecular structures. http://www.pdb.org/ |
| PubMed | Literature citations. http://www.ncbi.nlm.nih.gov/pubmed |
| Swiss-Prot/UniProt (23) | Link to the protein entry. Protein family, localization and catalysed reaction. http://www.uniprot.org/ |
| Databases specific for B. Subtilis | |
| DBTBS (24) | Link to the corresponding operon entry. Operon structure and regulation. http://dbtbs.hgc.jp/ |
| SubtiList (18)/GenoList (25) | Link to the corresponding gene entry. Name, length, product, function and genomic context of the genes. Molecular weight, isoelectric point and length of the protein. http://genolist.pasteur.fr/ |
| Database (reference) . | Extracted information, URL . |
|---|---|
| General databases | |
| EMBL-bank (16) | Link to the nucleotide and protein sequences. EC Numbers of enzymes. http://www.ebi.ac.uk/embl/ |
| KEGG (20) | Link to the corresponding gene entry. http://www.genome.jp/kegg/ |
| MPIDB (21) | Protein–protein interactions. http://www.jcvi.org/mpidb |
| PDB (22) | Link to the molecular structures. http://www.pdb.org/ |
| PubMed | Literature citations. http://www.ncbi.nlm.nih.gov/pubmed |
| Swiss-Prot/UniProt (23) | Link to the protein entry. Protein family, localization and catalysed reaction. http://www.uniprot.org/ |
| Databases specific for B. Subtilis | |
| DBTBS (24) | Link to the corresponding operon entry. Operon structure and regulation. http://dbtbs.hgc.jp/ |
| SubtiList (18)/GenoList (25) | Link to the corresponding gene entry. Name, length, product, function and genomic context of the genes. Molecular weight, isoelectric point and length of the protein. http://genolist.pasteur.fr/ |
Key publications used to seed SubtiWiki
| Extracted information . | References . |
|---|---|
| Essential genes | (26–28) |
| Gene regulation | (29–33) |
| Localization | (34–36) |
| Protein phosphorylation | (37–39) |
| Groups of related genes/proteins | (40–43) |
Key publications used to seed SubtiWiki
| Extracted information . | References . |
|---|---|
| Essential genes | (26–28) |
| Gene regulation | (29–33) |
| Localization | (34–36) |
| Protein phosphorylation | (37–39) |
| Groups of related genes/proteins | (40–43) |
The pages were generated using SubtiList, the semi-official database on B. subtilis (18). Moreover, information on gene products and functions, key quantitative properties of the genes (size) and proteins (size, molecular weight, isoelectric point) as well as the figure showing the genetic context were derived from SubtiList. The gene nomenclature used in SubtiList served as the basis for SubtiWiki. Gene designations that have been replaced since the last update in 2001 are still recognized with the SubtiList designation due to page redirects (see above). Information on protein families, the biological activities of proteins and their localization was, in addition to specific references, derived from SwissProt. The DBTBS database on transcriptional regulation in B. subtilis was used to extract information on transcription units, sigma factors and transcription regulators. The other databases provided important specialized information for many genes (Table 1).
Table 2 lists the articles that were of special importance for filling SubtiWiki with information. For this purpose, we selected publications that followed genome-wide strategies and that are therefore relevant to a large number of genes.
The data processing workflow
The databases and publications that served as source of information are very heterogeneous in the way of presenting information and in the use of gene designations. Data processing from each source consisted of five steps: (i) the acquisition of a holistic data set, (ii) mapping of the data to the gene designations in SubtiWiki, (iii) re-formatting the information to wiki markup language, (iv) batch upload of the information and (v) manual curation of the uploaded information. For all these steps, special purpose Python (www.python.org) scripts were developed.
Data were derived from either manually curated lists or from lists that were provided in the databases or publications. From these lists, the identifiers were parsed. If the identifiers were gene names, they were checked and, if necessary, converted to the actual annotation in SubtiWiki. For this purpose, a list of gene designations and synonyms (including redirects) was used. If accession numbers were used as identifiers we used tables that mapped the accession numbers to the SubtiWiki designations. The information was then re-formatted. The resulting entries conformed to the formatting style of MediaWiki and were enriched by internal and external links. For the upload, the Python scripts added the new information taking care not to replace any prior information. Since any automated process is prone to errors, some entries (about 1% of the entries) were randomly sampled to verify the correctness of the upload. Occasional systematic errors were corrected by new scripts or manually, if they were relevant only for a small number of pages.
Considerations for the attraction of a wide and active audience
The ultimate goal of research on any organism is to get a comprehensive understanding of its biology. A main part of this research is the elucidation of the functions of all genes, RNAs and proteins, their regulation, localization and interactions. By making such information available and easily accessible, SubtiWiki serves the scientific community that works on B. subtilis and closely related bacteria such as the pathogens S. aureus, L. monocytogenes or B. anthracis. SubtiWiki helps these communities to keep up with the research progress and provides insights into novel links between genes or proteins that might otherwise have escaped the attention of the busy scientist.
One could also imagine concentrating curation efforts on a small number of ‘large’ wikis instead of creating novel, more specialized wikis. Arguments in favour of the ‘central solution’ are the possibility of using the same set of tools and to provide a uniform interface for comparative genomics approaches. Nonetheless, there are also arguments against this approach: first of all, each model organism has its specific traits. The wiki should be tailored to the needs of the community that studies these traits, and the ‘one size fits all’ solution might be too restrictive. More importantly, as the use of scientific wikis is in an early stage, we are still in the process of identifying the best concepts that define a successful wiki. A healthy competition between alternative solutions will allow us to discern what works best.
To make SubtiWiki a vivid platform of information exchange, it needs the attention and the commitment of the Bacillus scientific community. With the information already provided for all genes (Figure 2), we are confident that any researcher looking at it will find it useful and we count on the willingness of the colleagues to share their expertise and to improve SubtiWiki by contributing. We trust that we provide already so much in terms of information content and simplicity of the interface that the hurdle is set quite low for any other potential contributor. This might be the solution for the problem that many scientific wikis experience, that is, the difficulty to attract new contributors (19).
In principle, there are two ways for finding information, the ‘ants’ perspective’ and the ‘birds’ perspective’. Wikis provide information usually in the ants’ perspective. However, SubtiWiki is complemented by a presentation of metabolic and regulatory pathways in B. subtilis (i.e. SubtiPathways). This presentation provides an approach to information on B. subtilis metabolism from the bird's eyes view. On the one hand, it allows visual navigation in the pathways and links the genes and enzymes to SubtiWiki. On the other hand, SubtiWiki provides links to SubtiPathways to visualise all genes/RNAs/proteins in the context of their cellular functions. The CellDesigner (44) source files of SubtiPathways (in Systems biology markup language) are available upon request.
At this early stage of using wikis in biocuration, we feel that SubtiWiki might prove to be useful for many scientists who are considering the creation of a scientific wiki for their own purpose.
Funding
Federal Ministry of Education [Research SYSMO network (PtJ-BIO/0313978D)] and the Fonds der Chemischen Industrie to J.S. and International Molecular Biology Program of the University of Göttingen and the Studienförderwerk Klaus Murmann der Stiftung der Deutschen Wirtschaft to L.A.F.
Conflict of interest statement: None declared.
Acknowledgements
The authors are grateful to Leendert Hamoen and Ulrike Mäder for helpful discussions.